-
PDF
- Split View
-
Views
-
Cite
Cite
Stefan Maas, Willemijn M. Gommans, Identification of a selective nuclear import signal in adenosine deaminases acting on RNA, Nucleic Acids Research, Volume 37, Issue 17, 1 September 2009, Pages 5822–5829, https://doi.org/10.1093/nar/gkp599
Close - Share Icon Share
Abstract
The adenosine deaminases acting on RNA (ADARs) comprise a family of RNA editing enzymes that selectively modify single codons within RNA primary transcripts with often profound impact on protein function. Little is known about the mechanisms that regulate nuclear RNA editing activity. Editing levels show cell-type specific and developmental modulation that does not strictly coincide with observed expression levels of ADARs. Here, we provide evidence for a molecular mechanism that might control nuclear import of specific ADARs and, in turn, nuclear RNA editing. We identify an in vivo ADAR3 interaction partner, importin alpha 1 (KPNA2) that specifically recognizes an arginine-rich ADAR3 sequence motif and show that it acts as a functional nuclear localization sequence. Furthermore, whereas KPNA2, but not KPNA1 or KNPA3, recognizes the ADAR3 NLS, we observe the converse binding specificity with ADAR2. Interestingly, alternative splicing of ADAR2 pre-mRNA introduces an ADAR3-like NLS that alters the interaction profile with the importins. Thus, in vivo RNA editing might be regulated, in part, through controlled subcellular localization of ADARs, which in turn is governed by the coordinated local expression of importin α proteins and ADAR protein variants.
INTRODUCTION
Nuclear pre-mRNA editing has been recognized as an important mechanism for the generation of RNA and protein diversity (1–3). In A-to-I editing, single adenosines in primary transcripts undergo deamination mediated by substrate-specific RNA fold-back structures and an adenosine deaminase acting on RNA (ADAR) (1). Editing can alter mRNA codons causing an amino acid substitution in the resulting protein with substantial functional consequences (2). In addition, A-to-I editing may create or abolish a pre-mRNA splice site (4,5) and high-level editing in repetitive element sequences may lead to nuclear retention of mRNAs (6,7). Furthermore, editing of miRNA precursors has been shown to alter biogenesis of miRNAs or the miRNA targeting profile (8). Importantly, human disease phenotypes have been linked to over- or under-editing of genes (9) and mouse models of editing deficiency or misregulation displayed profound phenotypes, such as embryonic lethality [ADAR1 knock-out (10,11)], epilepsy [ADAR2 knock-out (12)] and obesity [ADAR2 overexpression (13)].
It is currently not known what causes disease-related alterations in RNA editing levels. However, there is strong evidence that intracellular A-to-I RNA editing is a tightly regulated process. For example, the editing of several known substrates is subject to cell-type specific (14,15) and developmental regulation (16,17). In addition, it has been suggested that in some instances editing levels may change in response to external stimuli (18), but very little is known about the underlying regulatory mechanisms involved. Intriguingly, the observed changes in editing levels often do not correlate with the changes in the mRNA expression of editing enzymes (14,17,19).
Whereas in flies a single ADAR is responsible for all of the mRNA-directed A-to-I editing, in vertebrates a family of three ADARs has been characterized (20). ADAR1 and ADAR2 edit all currently known modification sites with distinct but overlapping substrate specificities. In contrast, the brain-specifically expressed ADAR3 protein has no documented deaminase activity (20) and there is neither an established function for the protein, nor any known physiological RNA target.
The overlapping and ubiquitous expression pattern of ADARs in mammalian tissues on one hand and the observed cell-type specific and ontogenetic regulation of editing levels on the other raise the question of how the editing activity of ADARs is regulated intracellularly.
Most known targets for editing constitute pre-mRNA molecules that need to encounter the ADAR protein in the nucleus before splicing, as intronic sequences are essential for forming the RNA fold-back structure recognized by ADARs. All ADARs harbor putative nuclear localization sequences (NLSs), but their function has been incompletely characterized. ADAR1 shuttles between nucleus and cytoplasm, in part, due to the presence of a nuclear export signal (21). Furthermore, ADAR1 was recently shown to be transported in and out of the nucleus through transportin-1 mediated binding to one of its dsRNA binding domains (22). In addition, within the nucleus, both ADAR1 and 2 have been shown to shuttle between the nucleoplasma and nucleoli (23,24).
The traditional nuclear import pathway involves the recognition of the classical nuclear localization sequence (cNLS) of the protein cargo by importin alpha, followed by importin beta binding, docking of the ternary complex with the nuclear pore complex (NPC) and transport of the complex across the nuclear membrane (25,26).
Nuclear transport of proteins is a highly regulated process, for example, by modulation of cNLS recognition through either post-translational modification of cNLS sequences or masking of the cNLS by heterologous molecules or alternative protein conformations. For instance, the nuclear activity of several transcription factors is determined through regulated nuclear import (27–29).
The nature of the individual cNLS sequence further impacts its recognition by importin alpha proteins. Monopartite cNLS signals are bound differently than bipartite NLS signals (30) further increasing the complexity of distinct importin alpha-cargo interactions. Importin alpha proteins consist of a N-terminal importin beta binding domain (IBB) and a domain composed of 10 tandem armadillo (ARM) repeats. The helical ARM repeats form the NLS binding sites and the 10th repeat binds the exportin CAS (31).
The importin alpha genes constitute a multi-gene family of three main subfamilies; importin alpha-S (human importin alpha 5, -6 and -7), alpha-P (human importin alpha 1) and alpha-Q (human importin alpha 4 and -3) (32). The importin alpha isoforms are expressed differentially based on northern blotting and western blotting studies (32–34). For example, human importin alpha 4 (KPNA3) makes up more than 1% of protein in skeletal muscle, but is essentially absent in spleen, kidney and heart (35). Recently, detailed in situ hybridization studies have shown regional specific expression of individual importin alpha subtypes in the brain (36,37). Whereas human importin alpha 5, 7, and 3 (KPNA1, 6, 4) are ubiquitously expressed in the brain, importin alpha 1 and 4 (KPNA 2, 3) show area-specific expression patterns.
Importin alpha proteins are believed to be involved in tissue-specific regulatory mechanisms (31). For example, in mouse embryonic stem cells, the switch from importin alpha 1 to alpha 5 can elicit neural differentiation (38). Importin-alpha-mediated nuclear import has also been shown to be involved in transporting synaptically generated signals into the nucleus during learning-type forms of plasticity (39). Currently, there are six importin alpha isoforms known in human with overlapping and distinct cNLS binding specificities (40).
In this study, we provide evidence that suggests a role for controlled nuclear import in RNA editing regulation. First, we screened for ADAR3 interaction partners using the unique ADAR3 N-terminal region as a bait in a yeast two-hybrid system. Importin alpha KPNA2 emerged as a protein that specifically recognizes the R-domain region within ADAR3, whereas KPNA1 and KPNA3 do not bind. Conversely, the main splice form of ADAR2 interacts with KPNA1 and KPNA3, but not KPNA2. Finally, an ADAR2 alternative splice form, ADAR2R, recently reported by us (41), displays an importin alpha interaction profile identical to the ADAR3 protein indicating that there is not only differential interaction between ADARs and importin alpha proteins, but that alternative splicing further modulates the nuclear import pathways for ADAR2.
MATERIALS AND METHODS
DNA constructs
For a list of oligonucleotide primer sequences used in this study see Supplementary Table S1.
The amino-terminal region of rat ADAR3 cDNA (42) including the R-domain (A3 1–133; 98% identical to human ADAR3) was generated by PCR with ADAR3 specific oligonucleotide primers and subcloned into the yeast expression vector pAS 2-1 (Clontech; Matchmaker Two-Hybrid System 2) which leads to expression of a fusion protein of the ADAR3 construct and the GAL4 DNA-binding domain. Additional ADAR3 constructs A3 1–220 (amino acids 1–220) and A3 220–390 were also generated by PCR and cloned into pAS 2-1. Point mutations into the R-domain of ADAR3 were introduced by site-directed mutagenesis (Stratagene site-directed mutagenesis kit) changing two of the Arginine residues to Serine.
Full-length rat ADAR2 cDNA (43) was subcloned into the yeast pAS 2-1 plasmid from a pBS II cDNA clone (kindly provided by Dr. Peter H. Seeburg, Heidelberg) by transferring first a Nco I/Nco I fragment (nt 1–462) and then a Nco I/EcoR I restriction fragment (nt 463–2140). Partial rat ADAR2 cDNA constructs (A2 1–110; A2 153–387; A2 387–701) were generated by PCR amplification and cloning of the cDNA into pAS2-1. The ADAR2R variant N-terminal cDNA construct A2R 1–112 was generated by PCR cloning using mouse brain cDNA template. Human ADAR1 (44) constructs A1 119–197 and A1 1–373 were constructed through PCR cloning (the human ADAR1 cDNA was kindly provided by Dr. Kazuko Nishikura, Philadelphia).
Full-length human KPNA1 (Accession BC002374) and KPNA3 (Accession NM_002267) cDNAs were generated by PCR using human brain random primed cDNA as a template and cloned into the pACT-2 yeast vector; the KPNA2 (Accession NM_002266) clone derived from two-hybrid screening also resides in plasmid pACT-2.
All final constructs of ADAR and importin proteins were subjected to DNA sequencing.
Two-hybrid interaction screen
The A3 1–133 ADAR3 N-terminal cDNA construct in pAS 2-1 was introduced into the Y190 yeast strain together with a human lung cDNA library (Clontech) subcloned into the pACT2 vector (Clontech) that fuses the library clones to the GAL4 transcriptional activation domain.
Selection of transformants on his–, leu–, trp– minimal medium allows growth only of colonies that harbor both expression plasmids and where the DNA-binding domain and activation domain of GAL4 are brought in close contact through an interaction between the ADAR3 protein and the library-derived protein sequence fused to the activation domain. Only then the His-promoter becomes activated. As a secondary test the β-galactosidase assay was used where the reconstituted GAL4 transcription factor activates a β-gal promoter. During selection, 15 mM concentration of the competitive yeast HIS3 protein inhibitor 3-AT (3-amino-1,2,4-triazole; Sigma) was used and the β-galactosidase assay will was performed on filter lifts of plates grown at 30°C for 2–4 days. After replica plating of the his-auxotroph colonies the yeast cells were permeabilized by one freeze/thaw cycle in liquid nitrogen and the β-galactosidase substrate X-Gal was applied in Z-buffer (Clontech). The filters were allowed to incubate for up to 12 h at 30°C.
The expression of the ADAR proteins in yeast was verified by western blot analysis using an HA-tag polyclonal antibody (Clontech) against the fusion protein.
In vitro transcription/translation and co-immunoprecipitation
The 35S-labeled and epitope-tagged ADAR and importin proteins were produced using an in vitro transcription/translation system (TNT coupled reticulocyte lysate system, Promega). For each protein, a PCR product was generated with the T7-RNA promoter consensus sequence on its 5′-end. The gel-purified DNA amplicons were used for RNA transcription and translation according to manufacturer's instructions using S35 methionine. Labeled proteins were analyzed using 10% polyacrylamide gel electrophoresis and autoradiography.
For co-immunoprecipitation, Protein A beads were washed twice with 500 μl 1× PBS, BSA was added to 1% (w/v) and preincubation of the beads at room temperature proceeded for 35 min. Beads were washed twice with 1× PBS, resuspended in Co-IP buffer [20 mM Tris–HCl (pH 7.5), 0.5% NP-40, 150 mM NaCl, 2 mM DTT], and added to the protein mixtures of antibody and labeled importin alpha and ADAR proteins. After incubation at room temperature for 1 h, beads were washed five times with Co-IP buffer, two times with 1× PBS and then resuspended with 20 μl SDS loading buffer. Fifty percent of each sample was run out on a 10% acrylamide gel, fixed, dried and exposed to autoradiography film.
Cell culture and immunofluorescence
The 14-amino-acid R-domain sequence of human ADAR3 was fused to the green fluorescent protein (GFP) by ligating an annealed pair of oligonucleotide primers that encode for amino acids 19–32 of ADAR3 into the pEGFP mammalian expression vector (Clontech) in frame to the vector encoded EGFP open reading frame.
HEK293 cells were cultured on glass coverslips under standard conditions and co-transfected using Polyfect transfection reagent (Gibco). After 24 h, the cells were rinsed in phosphate-buffered saline (PBS) solution, fixed for 30 min in 4% paraformaldehyde in PBS, and air dried. Expression of EGFP or EGFP–NLS fusion proteins was monitored using fluorescence microscopy (Leica DMRBE).
RESULTS
The R-domain of ADAR3 interacts with human importin KPNA2 in vivo and in vitro
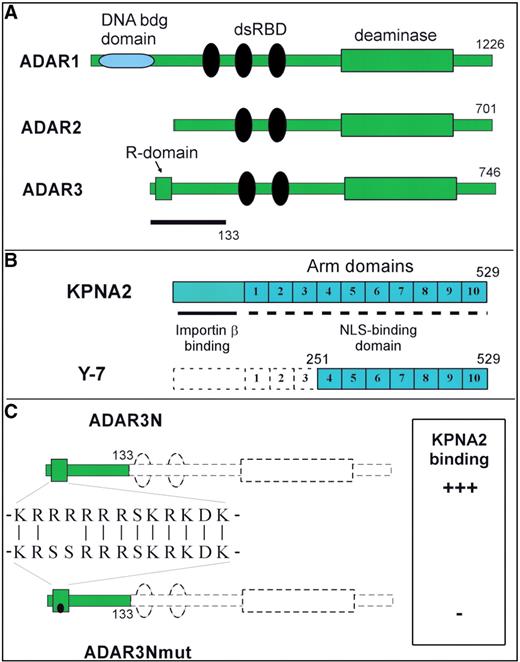
We performed a yeast two-hybrid interaction hunt (45) as a strategy to identify proteins that interact specifically with human ADAR3 protein, for which to date no specific function or editing target has been identified. As a bait for screening a human lung cDNA library, we used the distinctive N-terminal region of ADAR3 (42), located upstream of the first double-stranded RNA binding domain, fused to the GAL4 DNA binding domain (Figure 1A). This N-terminal region includes the arginine-rich R-domain of ADAR3, which previously had been shown to possess preferential binding affinity for single-stranded RNA (46). However, a specific target for binding or a functional role for ssRNA binding by ADAR3 have not been demonstrated.
(A) The ADAR3 construct used as bait in the yeast two-hybrid interaction hunt (amino acids 1–133) is depicted in relation to full-length ADAR3 (amino acids 1–746) and the domain structures of ADAR1 and ADAR2. ADAR1 refers to the human main splice form ADAR1a and ADAR2 corresponds to the human main splice form ADAR2a (57). dsRBD, double-stranded RNA-binding domain; deaminase, catalytic adenosine deaminase domain; R-domain, arginine-rich sequence motif. (B) The library clone Y-7, isolated from the two-hybrid screen as ADAR3-binding protein is outlined in relation to the full-length human importin α1 (KPNA2) sequence (Genbank Accession NM_002266). (C) The point mutant ADAR3Nmut, where two arginines within the R-domain have been replaced by serines, is shown in comparison to the ADAR3N construct. The binding activity to KPNA2 importin alpha according to the yeast interaction assay is indicated.
After screening a total of 3 × 106 independent yeast colonies we obtained one true positive his-auxotroph and β-galactosidase expressing colony after eliminating cases of self activation and other false positives.
Sequencing of the cDNA identified it as the importin α family protein KPNA2 (47,48), a receptor for NLSs. The isolated cDNA clone Y-7 (Figure 1B) encompasses amino acids 251–529 and therefore includes the ankyrin repeat motif (ARM) domains that have been shown to mediate the interaction of this NLS-receptor with its targets (49–51).
Next, we investigated if the R-domain of 11 amino acids located within the N-terminal region of ADAR3 is responsible for the observed interaction with KPNA2. The R-domain is characterized by a stretch of six consecutive arginines and several additional basic residues (Figure 1C). We introduced point mutations into the R-domain sequence by site-directed mutagenesis changing two of the arginine residues to serine (Figure 1C). The point mutant is expressed as a stable protein and in similar amounts as the wild-type N-terminal construct when introduced into a mammalian expression vector and expressed in HeLa cells according to western blot analysis (data not shown).
In the yeast two-hybrid assay, cells transformed with KPNA2 and this ADAR3 mutant (A3Nmut) do not grow on his–, leu–, trp– medium. Furthermore, the β-galactosidase reporter assay on leu–, trp– -grown colonies is also negative indicating that the mutation within the ADAR3 R-domain abolishes the interaction between the two proteins. Thus, the R-domain is required for the interaction of the N-terminal 133 amino acid construct of ADAR3 with KPNA2. When this 133-amino-acid ADAR3 construct (fused to a nonfunctional Gal4-DNA binding domain) is co-expressed in yeast together with the wild-type ADAR3/Gal4-DNA binding domain fusion and the KPNA/Gal4-activation domain fusion proteins, it competes with the productive interaction of the two fusion proteins as judged by a partial loss in galactosidase activity (data not shown). This observation further argues for a specific interaction between the 133-amino-acid ADAR3 sequence and KPNA2.
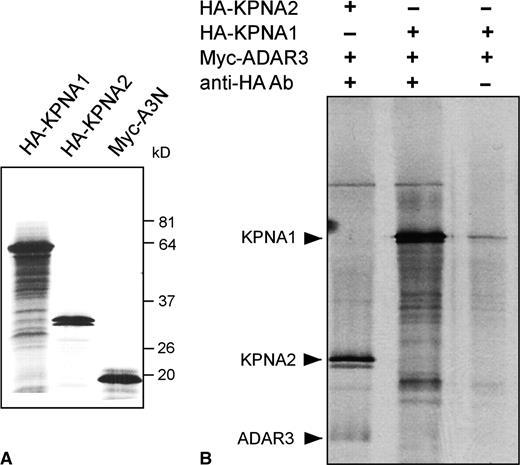
We then evaluated the in vivo interaction observed in the yeast two-hybrid system in an in vitro interaction assay (Figure 2). In co-immunoprecipitation experiments, S35-methionine labeled ADAR3 protein is specifically co-immunoprecipitated with a KPNA2-HA tagged protein when using a HA-tag specific monoclonal antibody. Interestingly, the observed in vitro binding between the ADAR3 N-terminal region and KPNA2 does not appear to be as robust as expected based on the clear-cut results in the yeast interaction assays. These differences may mirror distinct properties of in vitro transcribed and translated proteins versus in vivo expressed genes and could indicate the influence of post-translational modifications or effects of co-factors that modulate binding in vivo.
In vitro interaction of ADAR3N with KPNA2 but not KPNA1. (A) Expression of in vitro transcribed importin alpha proteins KPNA1 and KPNA2, as well as the ADAR3 protein A3N. Importin proteins carry the HA epitope tag, the ADAR protein a myc epitope tag. (B) Co-immunoprecipitation of importin/ADAR mixtures using a monoclonal anti-HA-tag antibody. In addition to KPNA2 protein, ADAR3N is co-immunoprecipitated (lane 1), but not when omitting the HA-tag specific antibody (lane 3). KPNA1 is not able to co-precipitate ADAR3N (lane 2).
The ADAR3 R-domain represents a functional cNLS and interacts specifically with KPNA2, but not with KPNA1 or 3
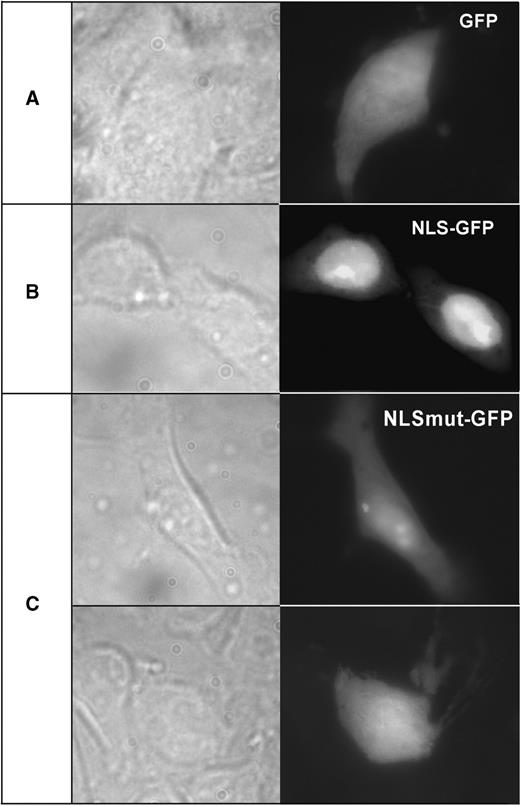
To determine if the R-domain sequence in ADAR3 constitutes a functional, classical NLS, we investigated if this isolated sequence motif is able to direct the nuclear import of the enhanced green fluorescent protein (EGFP) in living cells. To this end, we fused the 14-amino-acid R-domain sequence of human ADAR3 in frame to the EGFP cDNA. We then co-transfected into subconfluent human embryonic kidney cells (HEK 293) either the EGFP-R-domain fusion construct, the native EGFP plasmid lacking the ADAR3 sequence, or the EGFP-R-domain fusion construct with the previously described point mutation within the R-domain sequence and analyzed the subcellular distribution of the EGFP signal by fluorescence microscopy. As shown in Figure 3, EGFP alone is expressed in a diffuse manner in the cytoplasm and nucleus (Figure 3A), whereas the EGFP-R-domain fusion protein strongly accumulates within the nucleus (Figure 3B). Importantly, the fusion protein with a point-mutation in the ADAR3 R-domain, which abolishes binding to the importin alpha protein in the yeast two-hybrid assay, also looses the ability to target EGFP to the nucleus (Figure 3C).
The R-domain of ADAR3 confers nuclear localization on GFP protein. HEK293 cells cultured on glass coverslips and transfected with expression plasmids EGFP (A), EGFP-NLS (B) or EGFP-NLSmut (C) were fixed after 24 h and examined by fluorescent microscopy. The native EGFP protein was preferentially observed in the cytoplasm (A), whereas the EGFP-NLS fusion protein with the ADAR3 R-domain sequence accumulated in the nucleus with strongest staining in the nucleoli (B). The mutated NLS sequence prevents nuclear accumulation of EGFP with some staining still seen in the nucleoli (C).
We can therefore conclude that the ADAR3 R-domain sequence acts as a functional nuclear localization signal in vivo as it confers nuclear localization on a heterologous cytoplasmic protein.
Interestingly, when analyzing ADAR protein sequences using the PredictNLS server (52), the R-domain of ADAR3 is predicted with high likelihood to represent a nuclear localization signal (98.14% of proteins with that sequence are nuclear proteins, whereas 1.85% with such a sequence are known non-nuclear proteins) further supporting the experimental findings described above.
In human there are three subfamilies of importin alpha proteins and there have been reports of differential recognition of nuclear localization signals by the different subfamilies. KPNA2 is a representative of the importin alpha-P subfamily (36). In order to investigate if the ADAR3 R-domain NLS is recognized by other importin alpha family members, we generated yeast expression constructs encoding human representatives from each of the other two subfamilies; importin alpha-S and importin alpha-Q.
Intriguingly, when we tested these in the yeast two-hybrid assay with the ADAR3 N-terminal construct, both KPNA1 (importin alpha-S subfamily) and KPNA3 (importin alpha-Q subfamily) did not show interaction, while the importin alpha-P protein (KPNA2) again gave a strongly positive signal (Figure 4A). This surprising result lead us to test other ADAR protein constructs for their ability to interact with these importin alpha family members.
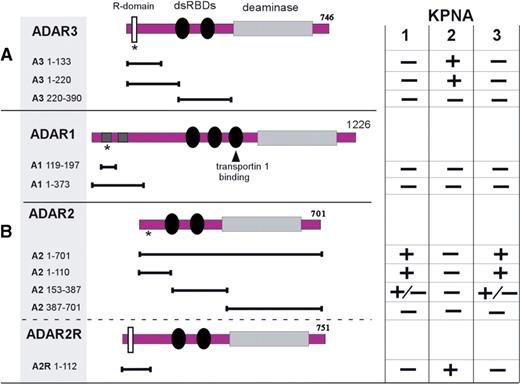
Interaction profiles of ADARs with importin alpha proteins. Depicted are ADAR3 (A) as well as ADAR 1, and 2 (B) expression constructs used in yeast two-hybrid interaction assays. Asterisks indicate the location of putative nuclear-localization signals; the dsRBD of ADAR1 that mediates transportin 1 shuttling (22) is indicated by an arrow. The binding activities of the listed constructs to KPNA1 to 3 importin alpha proteins according to a β-galactosidase assay on yeast colonies expressing pairs of proteins are indicated. In most cases, the results were either strongly positive (developing a blue color in less than 20 min) or clearly negative (no color after 24 h at 30°C). Occasionally, a weak signal developed after 12–24 h which is designated as ± in the table. dsRBDs: double-stranded RNA-binding domains; deaminase, catalytic adenosine deaminase domain; R-domain, arginine-rich sequence motif.
Differential interaction of importin alpha subfamily members with ADAR proteins and splice forms
We tested various combinations of ADARs and the three importin alpha proteins in the yeast two-hybrid assay. The ADAR constructs were designed to harbor the putative or known NLSs in ADAR1 and ADAR2. As summarized in Figure 4B, ADAR2 full-length, as well as shorter constructs that include a putative cNLS, display the opposite interaction profile as ADAR3. Whereas KPNA2 does not show interaction, both KPNA1 and KPNA3 give a strong positive signal in the galactosidase assay. The ADAR2 construct that encompasses only the C-terminal region including the catalytic domain (A2 387–701) did not interact with any of the KNPA proteins. A2 153–387, which includes the dsRNA binding domains of ADAR2, but lack the putative classical NLS, displayed weak but specific binding activity to KPNA1 and KPNA3 based on the galactosidase assay results. In contrast, the ADAR1-derived constructs lacked any detectable binding activity with the three importin alpha proteins. That result is not unexpected, since ADAR1 was recently shown to employ a distinct, nuclear transport pathway mediated by transportin-1 to shuttle between nucleus and cytoplasm, which involves a nonclassical NLS within the third dsRBD of ADAR1 (22).
We previously described a novel alternative splice form for ADAR2, which changes the open reading frame of the cDNA such that it extends the resulting protein by about 50 amino acids at its N-terminus (41). Intriguingly, we identified a sequence motif within this extension that is highly similar to the R-domain of ADAR3. We therefore hypothesized that the R-domain in this ADAR2 splice variant, termed ADAR2-R, may also constitute an NLS, which would convey an altered interaction profile with the importins compared to the major ADAR2 splice form lacking the N-terminal extension.
Indeed, when we test the ADAR2-R N-terminal sequence against the three importin alpha proteins, we see the same pattern of interaction as the one observed with the ADAR3 N-terminus (Figure 4B).
DISCUSSION
The ADAR3 R-domain was previously proposed to possess single-stranded RNA-binding activity (46). However, a functional role for such an activity has not been demonstrated. Furthermore, the ssRNA used in binding experiments was not a known or suspected target of ADAR3, but a randomly chosen sequence. The question if ssRNA binding is functionally relevant can only be answered in the context of a physiological RNA sequence or tertiary structure to which this ADAR would then bind, in part, through this R domain. However, without any known binding or modification target for ADAR3, this remains purely speculative.
In this study, we show that this R-domain specifically mediates interaction with a cellular nuclear localization signal receptor, and as an isolated sequence motif, can redirect GFP protein to the nucleus. The R-domain in ADAR3 is the only candidate cNLS sequence predicted by computational algorithms based on known classic NLS motifs (52). However, we cannot formally rule out, that there may be additional, nonclassical type import signals present in ADAR3.
Interestingly, it has recently been shown that the nuclear shuttling of the editing enzyme ADAR1 is co-regulated by RNA (22). In that case, an entire dsRNA binding domain (dsRBD) overlaps with the protein interaction surface bound by transportin 1. In fact, co-regulation of dsRBD-mediated nuclear export by dsRNA is well established (see citation 22 and references therein). Could the ssRNA-binding properties of the ADAR3 R-domain and its function as NLS indicate a somewhat similar mechanism? That may be unlikely. Experimentally, it would be difficult to test in vivo, if ssRNA binding influences protein interaction since due to the small size of the R-domain, the RNA and protein-binding cannot be dissociated through mutagenesis as was possible in case of the dsRBD of ADAR1 (22). Even if the observed in vitro affinity for ssRNA in general would be able to interfere with importin alpha binding also in vivo, and assuming that any type of physiologically relevant ssRNA concentrations could be reproduced experimentally, a biological function of this ADAR3 regulation by ssRNA binding is difficult to imagine. That would be different, if there were a known physiological RNA target for ADAR3 binding or/and modification. In that case, the interaction of ADAR3 with that target, mainly via its two dsRBDs, could be assisted and maybe enhanced through the interaction of the R-domain with a specific area of ssRNA present within that target. Under those circumstances, the general in vitro ssRNA-binding activity of the ADAR3 R-domain would indeed be relevant for editing function, but may be less so in the context of nuclear transport.
We demonstrate differential interactions between ADAR1-3 proteins and the importin alpha gene family. This suggests that the interplay between the expression patterns of ADARs and importins may determine what kind of nuclear A-to-I RNA editing will be enabled in a particular cell type. For example, turning on the expression of an importin alpha protein may induce nuclear import of specific ADAR proteins or splice variants leading to an increase in nuclear RNA-editing activity.
ADAR3 has not exhibited enzymatic editing activity to date, but has been implicated as a potential regulator of ADAR1 and ADAR2 activity (42,46,53). Therefore, the KPNA2-regulated nuclear import of ADAR3 could affect the regulation of ADAR1- or ADAR2-specific editing events. For example, the expression of KPNA1 or KPNA3 in the same cells would first determine if ADAR2 will be imported into the nucleus. Then, additional expression of KPNA2 would allow for ADAR3 co-regulation to modulate editing activity.
At this point, due to the paucity of either a known function for ADAR3 or any known RNA target, it is not possible to directly assess the impact of KPNA2-mediated specific nuclear import of ADAR3 on the function of this putative deaminase.
There have previously been reports suggesting differential interaction and nuclear transport of cargo molecules by members of the importin alpha family (54,55). For example, the inducible transcription factor STAT1 is recognized by human KPNA2, but not by KPNA1 (50).
In addition, the tissue or cell-type specific expression of importin alpha isoforms has been shown to control important biological processes by directing the nuclear import efficiency for a given cargo. For example, Drosophila importin alpha 3 (homolog to human KPNA4) specifically transports heat-shock factor dHSF into the nucleus. The lack of expression of importin alpha 3 during early embryogenesis prevents nuclear entry of dHSF until expression of importin alpha 3 is induced later in development (56).
Several recent studies have shed light on the differential expression patterns of importin alpha mRNAs and proteins. For example, within the human brain importin alphas are predominantly expressed in neurons, but only to low levels in glia (36). KPNA2 mRNA was expressed at low to moderate levels throughout the brain and spinal cord, but more intense signals were seen in limited regions, including the olfactory bulb and reticular system. In contrast, KPNA3 expression showed low or a lack of signal in the olfactory bulb, thalamus, cerebellum and spinal cord (36).
The expression of ADAR proteins and splice forms has not been investigated thoroughly on the protein level, but in situ hybridization studies, quantitative real-time PCR, northern blotting and microarray experiments show that ADAR1 and 2, although ubiquitously expressed across cell types, are present to different amounts in individual tissues and cell types and also subject to alternative splicing, and in case of ADAR2, RNA editing. ADAR3 expression is restricted to the brain with regional and cell-type-dependent expression levels (42). Overall, judging by expression survey data, such as the Allen mouse brain atlas (37), importin alpha and ADAR proteins show partially overlapping, but distinct mRNA expression profiles. However, the low spatial resolution of these expression studies currently precludes a more exact and unambiguous analysis.
Interestingly, the nuclear transport of ADAR1 seems to be mediated through nonclassical import pathways (22), whereas ADAR2 and ADAR3 exhibit classical NLS sequences that are recognized by importin alpha proteins. The alternative splicing of ADAR2 further contributes to the modulation of ADAR2 nuclear transport as it allows for the production of ADAR2 enzymes that differ in their binding profile with importins. The expression level of ADAR2R relative to the major splice form is low (<5%), except for hippocampus where it reaches ∼10% as judged by tissue-specific quantitative mRNA expression analysis (41).
In addition to the regulation of transport into the nucleus, it has been suggested that nuclear ADAR activity may be regulated through shuttling of both ADAR1 and ADAR2 between nucleolus and nucleoplasma (23,24). Furthermore, ADAR2 appears to harbor an additional noncanonical NLS that can confer nuclear localization in the absence of the putative classical NLS in its N-terminus (23).
Since RNA editing in most recoding cases is an obligatory nuclear event, the control of ADAR localization may be an important regulatory process. Eventually, only single-cell-level analysis of importin alpha and ADAR protein variant expression will be able to determine to what extent the nuclear import of RNA editing enzymes may be dynamically regulated based on interaction with KPNA family members in vivo.
Our observation regarding the apparent differences in the robustness of in vivo versus in vitro binding activities could be an indication that post-translational modification of basic residues directly within the R-domain may affect in vivo interaction. In future studies it may be investigated if the potential regulation of ADAR nuclear import is in turn regulated, for example, through methylation of arginine (58) residues or sumoylation of lysine residues (59) within the R-domain.
FUNDING
The National Institutes of Health (grant number NS057739 to S.M.). Funding for open access charge: Lehigh University.
Conflict of interest statement. None declared.
ACKNOWLEDGEMENTS
We thank Dr Kazuko Nishikura for providing ADAR1 cDNA and Dr Miyoko Higuchi and Dr Peter H. Seeburg for providing ADAR2 cDNA.




Comments