-
PDF
- Split View
-
Views
-
Cite
Cite
Mariona Nadal-Ribelles, Glòria Mas, Gonzalo Millán-Zambrano, Carme Solé, Gustav Ammerer, Sebastián Chávez, Francesc Posas, Eulàlia de Nadal, H3K4 monomethylation dictates nucleosome dynamics and chromatin remodeling at stress-responsive genes, Nucleic Acids Research, Volume 43, Issue 10, 26 May 2015, Pages 4937–4949, https://doi.org/10.1093/nar/gkv220
Close - Share Icon Share
Abstract
Chromatin remodeling is essential for proper adaptation to extracellular stimuli. The p38-related Hog1 SAPK is an important regulator of transcription that mediates chromatin remodeling upon stress. Hog1 targets the RSC chromatin remodeling complex to stress-responsive genes and rsc deficient cells display reduced induction of gene expression. Here we show that the absence of H3K4 methylation, either achieved by deletion of the SET1 methyltransferase or by amino acid substitution of H3K4, bypasses the requirement of RSC for stress-responsive gene expression. Monomethylation of H3K4 is specifically inhibiting RSC-independent chromatin remodeling and thus, it prevents osmostress-induced gene expression. The absence of H3K4 monomethylation permits that the association of alternative remodelers with stress-responsive genes and the Swr1 complex (SWR-C) is instrumental in the induction of gene expression upon stress. Accordingly, the absence of SWR-C or histone H2A.Z results in compromised chromatin remodeling and impaired gene expression in the absence of RSC and H3K4 methylation. These results indicate that expression of stress-responsive genes is controlled by two remodeling mechanisms: RSC in the presence of monomethylated H3K4, and SWR-C in the absence of H3K4 monomethylation. Our findings point to a novel role for H3K4 monomethylation in dictating the specificity of chromatin remodeling, adding an extra layer of regulation to the transcriptional stress response.
INTRODUCTION
The dynamics of chromatin structure are tightly regulated during transcription. Strategies to locally reconfigure chromatin structure include the use of adenosine triphosphate (ATP)-dependent chromatin remodeling complexes, the incorporation of histone variants within nucleosomes and the covalent modification of histones. ATP-dependent chromatin remodeling complexes play an essential role in the regulation of chromatin by altering the structure, composition and positioning of nucleosomes (1,2). Chromatin remodelers of the SWI/SNF subfamily such as the RSC complex establish a nucleosome-depleted region around the promoter, thus exposing it to the transcriptional machinery (3). Other complexes such as SWR1, a member of the INO80 subfamily, catalyze the ATP-dependent nucleosomal exchange of the canonical histone H2A with H2A.Z in nucleosomes flanking the nucleosome-free region. Subsequently, H2A.Z facilitates gene expression by creating unstable nucleosomes, through chromatin folding, and by recruitment of the transcription machinery (4–7).
In eukaryotic cells, Stress-Activated Protein Kinases (SAPK) play an essential role in proper cell adaptation to extracellular stimuli (8–11). Exposure of cells to high osmolarity results in rapid activation of a conserved family of SAPKs, which include mammalian p38 and yeast Hog1 (12). Hog1 is a key element in the reprogramming of gene expression in response to stress by acting on hundreds of stress-responsive genes (11,13–16). Hog1 is recruited to osmo-responsive promoters by specific transcription factors (17) and serves as a platform for the recruitment of RNA Pol II (18,19) and associated factors (20–22). Binding of Hog1 to chromatin also extends to the coding regions of stress-responsive genes where Hog1 associates with elongating RNA Pol II (19,23–25). The RSC complex is a chromatin remodeling complex that targets several stress-dependent genes. External stimuli induce changes in RSC genome-wide occupancy that correlate with the induction or repression of specific families of genes that are regulated by stress (26–28). In response to osmostress, Hog1 targets the RSC complex to coding regions to achieve efficient nucleosome eviction (28). The Ino80 chromatin remodeling complex appears to be important for the reassembly of chromatin during adaptation (29). The dynamic patterns of nucleosome positioning are central for establishing a threshold for gene induction upon Hog1 activation (30).
Here, we investigated the regulation of chromatin remodeling by the HOG signaling pathways, which is activated in response to extracellular stimuli. We show that methylation of the histone H3K4 by the histone methyltransferase Set1 determines whether RSC or Swr1 complexes can remodel nucleosomes at stress-responsive loci upon osmostress. Our results point to a new function for H3K4 monomethylation in governing the selectivity of chromatin remodelers to stress responsive genes, providing fine-tuning to the transcriptional response.
MATERIALS AND METHODS
Yeast strains, oligonucleotides and growth conditions
Tables containing the description of strains and oligonucleotides used in this study are enclosed in Supplementary Material. The rsc9ts derivative strains were grown at 25ºC and shifted to non-permissive temperature (37ºC) for 2 h.
Chromatin immunoprecipitation
Chromatin immunoprecipitation was performed as described previously (31,32). Yeast cultures were grown to early log phase (OD600 = 0.6–1.0) and exposed to osmotic stress treatment (0.4 M NaCl) for the time specified in the figure legends. Galactose induction was performed by growing cells onto YP 2% raffinose and adding 2% glucose or galactose. Strains carrying the ADGEV construct that expressed the Gal4DBD-hER-VP16 (33) were grown with 100 nM 17β-estradiol (17β-E2) and were then shifted to YPD or YPD plus 17β-E2 for 5 h before being shifted to a non-permissive temperature (37ºC). For cross-linking, yeast cells were treated with 1% formaldehyde for 20 min at room temperature. Antibodies used in this study were rabbit polyclonal antibodies to histone H3, H4 and mono-, di- and trimethyl H3K4 (Abcam antibodies ab1791, ab10158, ab8895, ab7766 and ab8580 respectively) and anti-Rpb1 (8WG16, Covance). A monoclonal anti-HA antibody was used to immunoprecipitate HA. Primer mixes for polymerase chain reaction (PCR) were adjusted for balanced signals. The following primers, which were utilized for conventional and real-time PCR analysis of stress genes and of constitutively expressed genes, were as follows. The locations indicate the distance from the respective ATG initiation codon: STL1 (-372/-112 for the promoter; +402/+630 for the 5′ coding region), CTT1 (-452/-160 for the promoter; +422/+669 for the coding region), GAL1–10 (-537/-215) and TEL1 (a 490 bp region on the right arm of chromosome VI) was used as a loading control. Experiments were performed using three independent chromatin preparations, and quantitative PCR analysis was performed in real time using an Applied Biosystems 7700 sequence detector. Immunoprecipitation efficiencies were calculated in triplicate by dividing the amount of PCR product in the immunoprecipitated sample by the amount of PCR product in the input sample. Data are presented as fold immunoprecipitation over the TEL1 sequence control.
Northern blot analysis
Yeast strains were grown to mid-log phase as described for the ChIP experiments and were then subjected to osmotic shock (0.4 M NaCl) for the indicated times and conditions. Total RNA and expression of specific genes were probed using labeled PCR fragments containing the entire open reading frame of STL1 (1.7 kbp), CTT1 (1.7 kbp), ACT1 (1.4 kbp), RDN18 (1.8 kpb) and ENO1 (1.3 kbp).
MNase nucleosome mapping
Yeast spheroplast preparation and micrococcal nuclease digestions were performed as described previously with modifications (16). Spheroplasts were prepared from mid-log phase cultures grown in YPD before being subjected to osmostress (0.4 M NaCl, 10 min). The cells were cross-linked with 1% formaldehyde for 15 min, the reaction was stopped with 125 mM glycine for 5 min. Cells were washed and resupended in 1M sorbitol TE buffer before cell wall digestion with 100 T zymoliase (USB). Cells were then lysed and immediately digested with 7.5–125 mU of micrococcal nuclease. For naked DNA controls, genomic DNA was extracted as previously described and digested with 0.003–0.2 mU of micrococcal nuclease under the same conditions. Tiled primers covering the entire coding region and 500 bp upstream and downstream of the STL1 locus were used for nucleosome positioning (amplicons of 100 pb with an overlapping of 20 pb). Specific oligonucleotides are included in the Table provided in Supplementary Material. qPCR analysis was performed to determine nucleosome positioning and was normalized by naked DNA.
RESULTS
Deletion of SET1 or H3K4 mutation bypasses the requirement for RSC in osmostress gene expression
DNA is wound around proteins called histones in a structure termed chromatin. For gene expression, the DNA must be unwound so that proteins necessary for gene transcription can access the DNA. Thus, induction of strongly activated genes requires major chromatin reorganization (15). Chromatin remodeling at stress-responsive genes is a hallmark of successful gene induction upon osmostress (19). Here, chromatin remodeling is mediated by Hog1 and depends on the RSC chromatin remodeling complex (19,28).
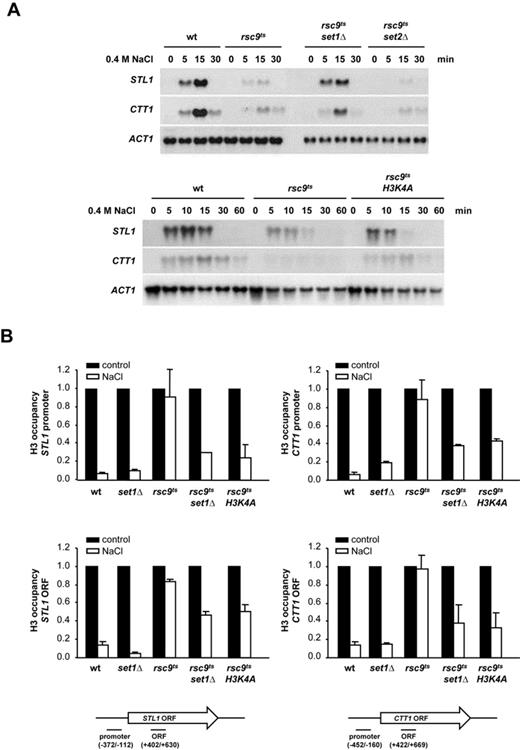
A search for suppressors of rsc9ts heat sensitivity provided a hint that there might be a connection between the RSC complex and histone methylation (J. Balog, R. Dechant and G. Ammerer, unpublished observation). To study this potential connection, we tested whether deletions in the SET1 or SET2 methyltransferases could suppress the transcriptional defect observed in the rsc9ts or rsc9 degron strains in response to high osmolarity. As shown in Figure 1A and Supplementary Figure S1A, deletion of SET1 suppressed the defects in osmostress responsive CTT1 and STL1 gene expression caused by RSC inactivation. In contrast, deletion of the histone methylase SET2 did not suppress the rsc9ts mediated defects in gene expression upon stress (Figure 1A). RSC is a multiprotein complex of several components. Of note, suppression of the RSC defect by set1Δ was not restricted to rsc9 mutants since we obtained similar gene expression results in rsc8deg set1Δ cells (not shown). It is worth noting that SET1 mutation did not suppress other phenotypes associated with RSC deficiency such as sensitivity to caffeine or heat-responsive gene expression (Supplementary Figure S1B and SC) indicating its specificity for the osmostress response.
Deletion of SET1 or H3K4A mutation partially suppresses the defects in osmostress gene expression and chromatin remodeling caused by rsc9ts mutation. (A) Deletion of SET1 and mutation of K4 in histone H3 restore gene expression in response to osmostress in an rsc9ts strain. The indicated strains were grown in YPD at 25ºC until OD660 0.5, shifted to non-permissive temperature (37ºC) for 2 h and subjected to osmostress (0.4 M NaCl) for the indicated times. Total RNA was assayed by northern blot for STL1, CTT1 and ACT1 (as a loading control) expression. (B) Deletion of SET1 and mutation of K4 in histone H3 partially suppress the defect in osmostress nucleosome eviction caused by rsc9 mutation. Wild type, rsc9ts, set1Δ, rsc9tsset1Δ and rsc9ts H3K4A mutant strains were treated as in (A) and histone H3 occupancy of the promoter or open reading frame (ORF) of STL1 or CTT1 loci was then analyzed before (filled bars) or after osmostress (open bars) using ChIP assay followed by real-time PCR. Histone H3 occupancy was normalized to internal telomere control. Data represent the mean and standard deviation of three independent experiments. Schematic representation of the STL1 and CTT1 locus depicting primer positions is shown. Detailed information about primer position and sequence is provided in Supplementary Table S2.
We have previously identified other transcriptional regulatory complexes for stress-responsive genes, such as SAGA and RPD3, which are required for induction upon osmostress (20,21). Thus, we then tested whether mutation of SET1 could suppress the defect in gene expression that it was associated with SAGA and RPD3 mutants. As shown in Supplementary Figure S2A, deletion of SET1 did not render significant changes in gene expression in the SAGA or RPD3 mutant strains. Thus, SET1 suppression is associated specifically with RSC and not with other transcriptional elements that are involved in Hog1-mediated transcription response.
The Set1 histone methylase is involved in the specific methylation of lysine 4 in histone H3 (H3K4) (34–36). To analyze whether the suppression of RSC by set1Δ was due to defective H3K4 methylation, we analyzed gene expression in an rsc9ts strain containing a mutation of histone H3 lysine 4 to alanine (H3K4A). As shown in Figure 1A, following osmostress, gene expression was increased in the rsc9ts H3K4A cells when compared to the rsc9ts cells. It is worth noting that neither the deletion of SET1 nor the H3K4A mutation alone altered the kinetics of expression of STL1 or CTT1 (Supplementary Figure S2B). Therefore, methylation of histone H3K4 prevents gene expression in the absence of RSC. These data suggest that methylation by Set1 imposes an extra layer of negative control on the expression of stress-responsive genes.
Deletion of SET1 or H3K4 mutation stimulates osmostress-induced chromatin remodeling in the absence of the RSC complex
Since RSC inactivation impairs loss of histone in stress-responsive in response to osmostress (28), we tested whether mutation of SET1 or H3K4A also suppressed such defect in the rsc9ts strain. Chromatin from wild type, rsc9ts, set1Δ, rsc9tsset1Δ or rsc9ts H3K4A cells subjected to osmostress was immunoprecipitated using specific antibodies against H3 and was then analyzed by PCR. The deficiency in H3 histone remodeling in the rsc9ts strain at STL1 and CTT1 loci following osmostress was partially reversed in cells containing SET1 or H3K4A mutations (Figure 1B). Therefore, the lack of histone H3K4 methylation bypasses the requirement for RSC to remodel chromatin in response to stress.
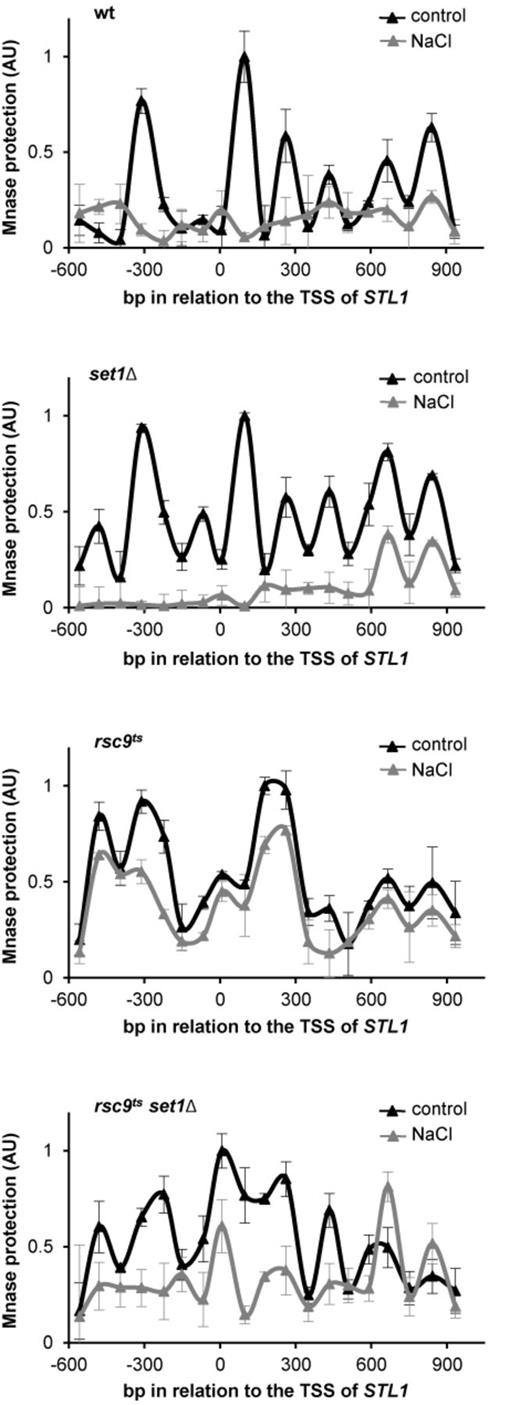
To analyze the role of histone methylation in nucleosome remodeling following osmostress, we then analyzed the nucleosome positioning at the STL1 locus by Micrococcal Nuclease (MNase) digestion of chromatin before and after stress in wild type, rsc9ts, set1Δ and rsc9tsset1Δ strains. As described, RSC action affects nucleosome position in basal conditions (37,38). We found that, as expected, inactivation of Rsc9 prevented the dramatic changes in chromatin structure observed upon stress in the wild type strain. Nevertheless, deletion of SET1 in the rsc9ts background partially restored such chromatin remodeling (Figure 2). Together, our data suggest that in cells deficient for the RSC complex, methylation by Set1 prevents chromatin reorganization upon stress.
Set1 prevents chromatin remodeling in the absence of RSC. Set1 mediated methylation of H3K4 inhibits chromatin remodeling in RSC mutants. The indicated strains were grown in YPD at 25ºC until OD660 0.5 and shifted to under non-permissive temperature (37ºC) for 2 h. Nucleosome positioning at chromosomal STL1 was assessed by MNase digestion of chromatin of wild type (wt), set1Δ, rsc9ts or rsc9tsset1Δ cells that were treated (gray triangles) or not (black triangles) with 0.4 M NaCl for 10 min. MNase digested DNA was isolated and analyzed using qPCR with tiled primers spanning the promoter and coding regions (-600 bp upstream and 900 bp downstream of the transcription start site (TSS), respectively). The normalized nucleosome occupancy is shown (x-axis) relative to the TSS. Data represent the mean and standard deviation of three independent experiments.
Stress-responsive genes are methylated by Set1 under non-stress conditions
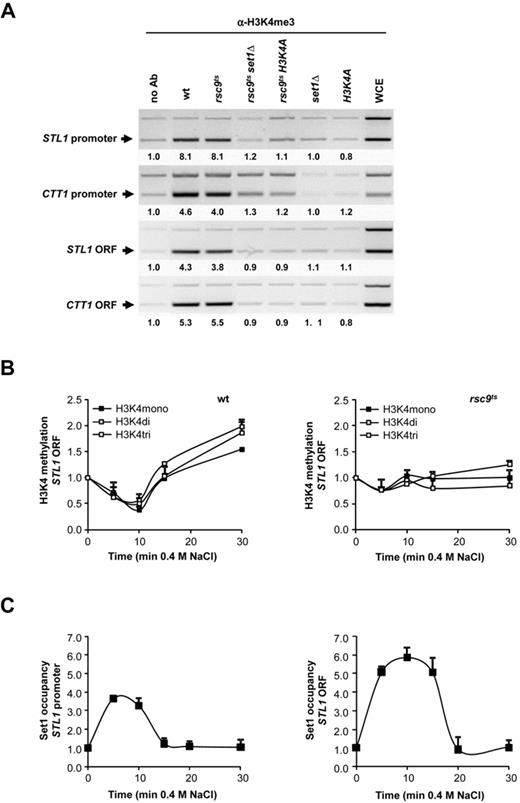
The suppression of the rsc9ts transcriptional defect by Set1 suggested the possibility that H3K4 methylation prevents chromatin remodeling and gene expression at stress-responsive loci under non-stress conditions. To assess whether histones at stress-responsive genes are methylated already in the absence of stress, we performed ChIP analysis in non-stressed cells, using antibodies against the H3K4 trimethylated form. Indeed, even in the absence of stress, trimethylated H3K4 (H3K4me3) was detected at the STL1 and CTT1 genes, both at the promoter and the ORF, in wild type cells or in rsc9ts strains (Figure 3A). In contrast, H3K4me3 was not detected in histone H3K4A or in set1Δ mutants. Notably, the level of H3K4 trimethylation observed at the STL1 locus under non-inducing conditions was comparable to that observed at the locus of an induced gene such as GAL1–10 in response to galactose (Supplementary Figure S3).
Trimethylation of osmostress genes is mediated by Set1. (A) Histone 3 is trimethylated at osmo-responsive genes under non-stress conditions. Wild type, rsc9ts, rsc9tsset1Δ, rsc9ts H3K4A, set1Δ and H3K4A strains were grown at 25ºC and shifted to under non-permissive temperature (37ºC) for 2 h until mid-log phase. Samples were taken for ChIP using a trimethyl H3K4 (Abcam antibody) to determine the levels of trimethyl H3K4 at the promoter and coding regions (ORF) of STL1 and CTT1 (same primers used in Figure 1B). As controls, chromatin from wild type cells was immunoprecipitated without antibody (no Ab) and DNA was amplified from extracts prior to immunoprecipitation (Whole-Cell Extract, WCE). PCR samples were amplified using TEL as a loading control (upper band). Quantification is depicted as fold increase over TEL. (B) Dynamics of H3K4 mono-, di- and trimethylation in the wild type and in the rsc9ts mutant cells in response to stress. Cells were grown at 25ºC and shifted to non-permissive temperature (37ºC) for 2 h until mid-log phase. Mono-, di- and trimethylation of H3K4 at the STL1 coding region was quantitatively analyzed using ChIP and real-time PCR in a wild type (left panel) and rsc9ts (right panel) strains after addition of 0.4 M NaCl for the indicated times. TEL was used as a reference control and levels of untreated samples were set to1 as a reference of each strain. Data represent the mean and standard deviation of three independent experiments. (C) Set1 is recruited to stress-responsive genes in response to osmostress. Binding of Set1-HA to the STL1 promoter and ORF was determined using ChIP and subsequent PCR analysis of Set1-HA tagged cells that were grown to mid-log phase and subjected to osmostress (0.4 M NaCl) for the indicated times. A telomere region was used as an internal control. Data represent the mean and standard deviation of three independent experiments.
Upon stress, removal of histones from a stress-responsive locus such as that of STL1 leads to a reduction in H3K4me3, but this trimethylation of H3K4 reappears as soon as nucleosomes are reassembled at the stress-responsive locus. Of note, histones are not evicted from the stress-responsive locus upon stress in an rsc9ts mutant and thus H3K4me3 levels remain constant (Figure 3B and Supplementary Figure S4). Corresponding with the redeposition of this H3K4 methylation, Set1 associates with stress-responsive genes such as the STL1 gene upon stress. At later time points, when maximal H3K4me3 has been redeposited, Set1 disassociates from the stress-responsive gene (Figure 3C). Thus, chromatin at stress-responsive genes is methylated at H3K4 both under basal conditions and after chromatin reassembly following stress.
H3K4 monomethylation inhibits gene expression of stress-responsive genes
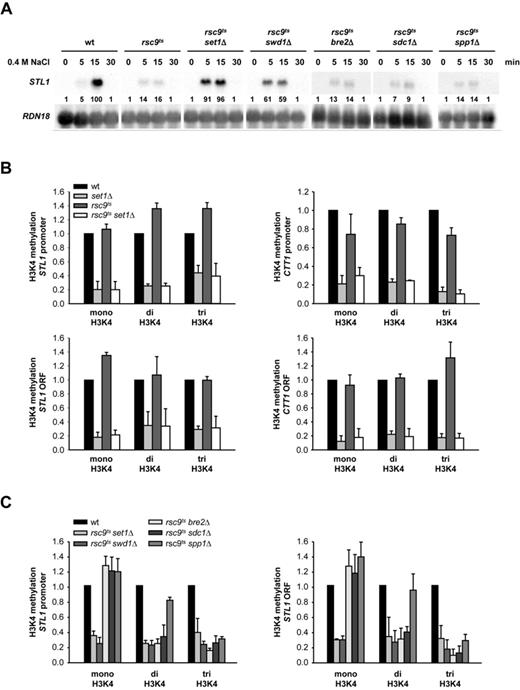
The Set1 complex mediates mono-, di- and trimethylation of H3K4 (36). We therefore sought to determine which modification of H3K4 might be responsible for regulation of the gene expression of stress-responsive genes. We first evaluated which degree of H3K4 methylation was relevant for osmostress gene expression by deleting individual Set1 complex subunits that specifically abrogate the levels of mono-, di- or trimethylation at H3K4. The Swd1 subunit is required for all three H3K4 methylation states, while Bre2 or Sdc1 can mediate both di- and trimethylation, and disruption of Spp1 only affects H3K4 trimethylation (39–41). We therefore mutated the SWD1, BRE2, SDC1 or SPP1 genes in an rsc9ts background and monitored gene expression upon stress. STL1 expression was not restored in rsc9ts cells containing bre2, sdc1 or spp1 mutations. In clear contrast, mutations in set1 and swd1 did suppress the RSC defect in transcription of STL1 (Figure 4A). These data indicate that the H3K4 monomethylation is the histone mark responsible of preventing gene expression of stress-responsive genes.
Monomethylation of H3K4 inhibits transcription. (A) Mutation of BRE2, SDC1 and SSP1 in an rsc9ts background strain does not bypass the transcriptional defect of rsc9ts mutation in osmostress gene expression. The indicated strains were shifted for 2 h to a non- permissive temperature before being subjected to osmostress (0.4 M NaCl) for the indicated times. Total RNA was assayed by northern blot for STL1 and RDN18 (as a loading control) expression. Quantification data came from the same original blot and it was normalized to the loading control. The value of maximum gene expression of the wild type strain was used as 100% reference. (B) Methylation state of H3K4 at osmo-responsive genes in the absence of stress. Cells were grown at 25ºC and shifted to non-permissive temperature (37ºC) for 2 h. Mono-, di- and trimethylated H3K4 at the STL1 and CTT1 promoters and ORFs under non-stress conditions were assessed using ChIP assays of wild type (black bars), set1Δ (light gray bars), rsc9ts (dark gray bars) and rsc9ts set1 (white bars) strains. Methylation levels were normalized to total H3 of the wild type strain whose levels were set to one and used as a reference for the mutant strains. (C) Mono-, di- and trimethylation of H3K4 at the STL1 promoter and ORF under non-stress conditions in wild type and in rsc9ts, rsc9tsset1Δ, rsc9tsswd1Δ, rsc9tsbre2Δ, rsc9tssdc1Δ and rsc9tsspp1Δ mutant strains was assessed using ChIP assays as in Figure 4B. Data represent means and standard deviation of three independent experiments.
We next analyzed the degree of H3K4 methylation associated with STL1 and CTT1 in wild type, set1Δ, rsc9ts and set1Δ rsc9ts cells in the absence of stress, using ChIP assays and specific antibodies against H3K4 mono-, di- and trimethylated antibodies. H3 was found to be mono-, di- and trimethylated at both STL1 and CTT1 promoters and coding regions (Figure 4B). As expected, H3K4 methylation was absent in strains carrying the set1 deletion. Of note, the methylation status of H3K4 in each Set1 complex mutant strain showed the expected pattern and only mutations in SET1 and SWD1 completely abolished all forms of H3K4 methylation (Figure 4C).
Overall, these data suggest that monomethylation of H3K4 is sufficient to determine the requirement for RSC in gene expression and chromatin remodeling. Of note, the levels of monomethylation in STL1 remained constant upon stress in an rsc9ts strain, in contrast to the reduction observed in wild type cells (Figure 3B).
The SWR1 complex and H2A.Z mediate chromatin remodeling and gene expression in the absence of RSC and H3K4 monomethylation
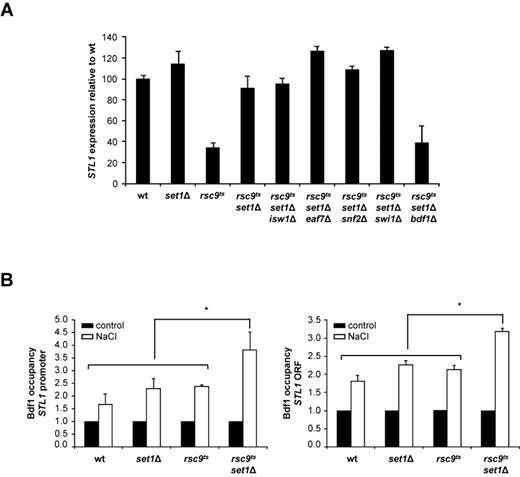
Since lack of H3K4 methylation suppressed the RSC deficiency and permitted nucleosome remodeling in response to stress, we hypothesized that an alternative remodeler might act at stress-responsive genes in the absence of H3K4 methylation. To identify such a chromatin remodeler(s), we performed a targeted candidate screen in which we deleted genes encoding representative members of all chromatin remodeling complexes in yeast (a total of 20 were assayed), in the set1 rsc9ts background. We reasoned that in the absence of this alternative remodeler the set1 deficiency should not suppress the rsc9ts mutation. When monitoring gene expression in these mutant strains, we found that only the absence of BDF1 in that background rendered cells that were unable to induce gene (STL1 and CTT1 in lesser extend) expression upon stress (Figures 5A and 6A). We then tested whether Bdf1 was recruited to stress-responsive genes upon osmostress, using ChIP assays. Although Bdf1 indeed associates with STL1 upon stress in wild type, set1Δ and rsc9ts strains, its binding was more robust in set1 rsc9ts deficient cells (Figure 5B). Of note, we did not detect differences in Bdf1 recruitment when comparing wild type, set1Δ, rsc9ts and set1Δ rsc9ts mutant strains in the absence of stress (Supplementary Figure S5).
Absence of methylation licenses the recruitment of other remodeling factors. (A) The absence of BDF1 in an rsc9ts set1Δ background impairs transcription of osmo-responsive genes. The indicated strains were shifted for 2 h to a non-permissive temperature before being subjected to osmostress. Total RNA was extracted from unstressed and stressed cells (0.4 M NaCl, 10 min) and the expression of STL1 was monitored by northern blot in the indicated strains. The graph represents the mRNA abundance of STL1 in response to osmostress, normalized to the loading control, using expression of STL1 from wild type as a reference. (B) Recruitment of Bdf1 to stress responsive genes is stimulated in response to osmostress in an rsc9ts set1Δ background. Association of Bdf1–6HA with the promoter and ORF of STL1 was analyzed in the indicated strains in the absence (black bars) or presence (10 min of 0.4 M NaCl, white bars) using ChIP assays. The real-time PCR results are shown as fold induction of treated relative to untreated (control) samples, which were normalized to a telomere internal control. Data are means and standard deviation of three independent experiments. (*) The statistical significance of Bdf1 recruitment upon stress was assessed by a paired T-student test of acceptance of equality at (P-value <0.05) comparing the wild type, set1Δ and rsc9ts versus rsc9ts set1Δ strains.
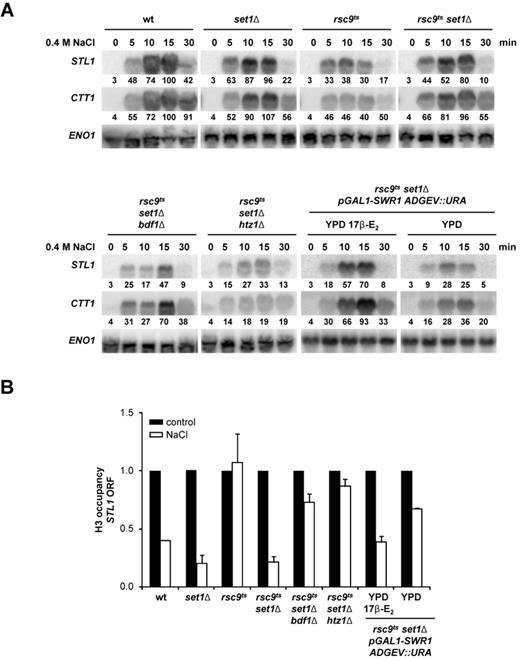
SWR1 is licensed to act as a chromatin remodeler in the absence of RSC and histone methylation. (A) Deletion of BDF1, HTZ1 and inducible expression of pGAL1::SWR1 prevents induction of osmo-responsive genes in an rsc9ts set1Δ background. The indicated strains were grown in either YPD or YPD 17β-estradiol (17 β-E2) before being shifted to a non-permissive temperature (37ºC) for 2 h. The cells were subsequently exposed to an osmotic stress of 0.4 M NaCl. Total RNA was assayed by northern blot for STL1, CTT1 and ENO1 (as loading control) expression. Quantification data came from the same original blot and it was normalized to the loading control. The value of maximum gene expression of the wild type strain was used as 100% reference. (B) Impairment of the SWR1 complex leads to defective chromatin remodeling. Total H3 eviction from the coding region of STL1 was followed using ChIP assays in the same strains and under the same conditions as in (A). The levels of total H3 under untreated (black bars) conditions were used as a reference for the treated samples (0.4 M NaCl, 10 min, white bars). Data represent the mean and standard deviation of three independent experiments.
Bdf1 is a component of the SWR1 complex that mediates H2A/H2A.Z histone exchange (4,42). To determine whether the Swr1 complex and H2A.Z exchange promote stress-induced gene expression and chromatin remodeling in the absence of RSC and H3K4 methylation, we assessed whether depletion of Swr1 or H2A.Z had the same effect as Bdf1 depletion. Since deletion of SWR1 in a set1 background results in slow growth, we introduced the SWR1 gene under the control of a GAL1 promoter using the pADGEV plasmid (33). This system allows to induce Swr1 expression from the GAL1 promoter by using estradiol. In the presence of estradiol, Swr1 was expressed and transcription of the stress-responsive genes STL1 and CTT1 was partially restored in a set1 rsc9ts strain. In clear contrast, induction of STL1 and CTT1 gene expression upon osmostress was impaired in the absence of Swr1 (Figure 6A). We then assessed whether the effect of Swr1 was mediated through H2A.Z. Deletion of the HTZ1 gene that encodes for H2A.Z in the set1 rsc9ts background also prevented STL1 and CTT1 gene expression upon stress (Figure 6A). Consistent with these data, in the set1 rsc9ts background, the lack of BDF1, SWR1 or HTZ1 impaired chromatin remodeling at the STL1 locus, as assessed by ChIP analysis of H3 occupancy of the STL1 ORF (Figure 6B). Taken together, our data suggest that the Swr1 complex and its effect on H2A/H2A.Z exchange promote gene expression and chromatin remodeling in the absence of RSC and H3K4 methylation.
DISCUSSION
Modifications at the surface of nucleosomes are critical for the regulation of chromatin dynamics and transcription (43). Histone methylation is one of those modifications that play a key role in the regulation of gene expression. Here, we found that methylation of H3K4 dictates the specificity for the complex that remodels chromatin at osmostress-responsive genes.
In the presence of H3K4 methylation, stress-responsive genes are remodeled by the RSC complex thereby permitting gene induction upon stress. Correspondingly, the absence of RSC largely impairs chromatin remodeling and gene expression at stress-responsive genes (28). Surprisingly, suppression of H3K4 methylation by deleting SET1 or mutating H3K4 to Ala, resulted in an increase in gene expression and chromatin remodeling in rsc deficient cells. These results suggest a negative role for H3K4 methylation by Set1 in gene expression. Set1 activity is most commonly associated with genes that are actively transcribed (44). However, data obtained from genome-wide analyses of the effects of H3K4 methylation are controversial; while some studies propose a global reduction in transcription in the absence of H3K4 methylation (45,46), others suggest that H3K4 methylation may play a crucial role in transcriptional repression (47,48). Examples for the role of Set1 and H3K4 methylation as a repressive mark are the ribosomal gene repression that occurs in response to diamide (49) or the repression of inducible genes in diverse mammalian cell types (50). Moreover, removal of H3K4 trimethylation has repressive effects on a specific subset of genes through promotion of 3′-end antisense transcription or the process on antisense transcription itself (51–54).
Here, we found that the presence of H3K4 methylation prevents gene transcription upon stress in the absence of RSC. Indeed, the use of mutants of specific genes, whose activities are responsible for the mono-, di- or trimethylation of H3K4, indicated that even monomethylation of H3K4 is sufficient to prevent gene transcription upon stress. After stress, nucleosomes are reassembled at stress genes (29). However, in principle those nucleosomes should be unmethylated and thus, without the monomethylation mark. Of note, Set1 is recruited to stress-responsive genes after stress and an increase in the methylation of newly incorporated nucleosomes is known to occur with similar dynamics as chromatin reassembly after stress (28,30). Thus, Set1 may act as a safeguard mechanism to maintain methylated chromatin at stress-responsive genes.
The fact that the absence of monomethylation of H3K4 allows chromatin remodeling and gene expression upon osmostress in rsc deficient cells led us to hypothesize that histone H3K4 methylation may impose RSC-dependent remodeling. In other words, in the absence of H3K4 methylation, alternative chromatin remodelers may gain access to stress-responsive genes to promote remodeling upon stress. Systematic analysis of different chromatin remodelers identified Bdf1, a subunit of the SWR1 complex, as an essential element to induce chromatin remodeling and gene expression upon osmostress in the absence of RSC and H3K4 methylation. Consistent with this finding, Bdf1 was recruited to stress responsive genes upon stress. The SWR1 complex controls H2A.Z dynamics (5–7,55). We found that the absence of the Swr1 catalytic subunit of the complex or of H2A.Z compromised chromatin remodeling and impaired gene expression upon osmostress. These data suggest a model by which H3K4 methylation specifies the requirement for a specific chromatin remodeling machinery, and that the lack of H3K4 methylation allows association of the Swr1 complex with osmostress-responsive genes to mediate chromatin remodeling and gene expression. Albeit the main effect of SWR1 is seen in the absence of RSC and H3K4me and, thus RSC is the major remodeler of stress-induced genes especially when histones are methylated, there is the possibility that not all histones are methylated under non-stress conditions and thus, SWR1 complex can act there even in the presence of RSC.
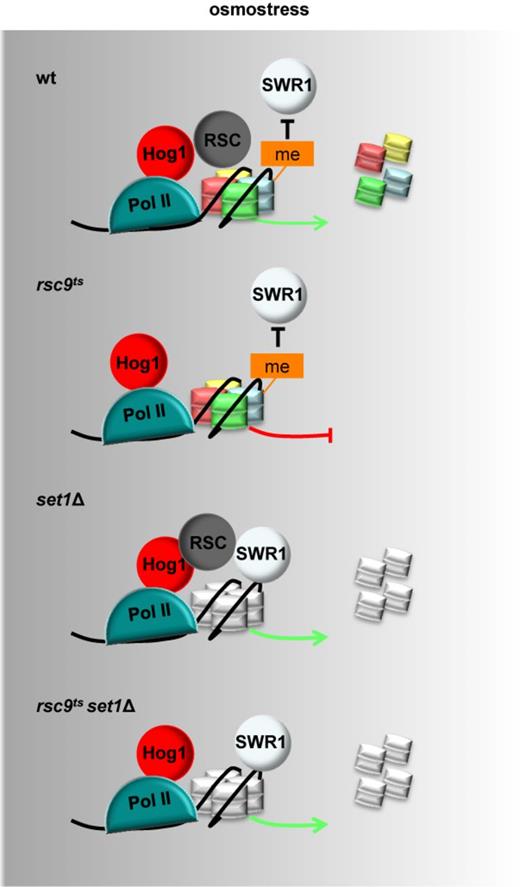
Our data outline a novel role for H3K4 monomethylation in dictating the specificity for chromatin remodeling, thus acting as an additional layer of transcriptional control (Figure 7). In this scenario, H3K4 methylation serves to define two states of chromatin. In the first state, when methylation is present at H3K4, then RSC is the chromatin remodeler that has to access stress-responsive genes to modify chromatin and to induce gene expression. It has been shown that activation of stress-responsive genes leads to strong nucleosome reorganization (19) and thus, deficient chromatin remodeling leads to impaired gene expression. Surprisingly, RSC activity is dispensable in the absence of methylation. It is then, under this second condition, that the Swr1 complex promotes chromatin remodeling via H2A.Z. It has been shown that nucleosomes with associated H2A.Z display more dynamic properties that facilitate gene expression (55,56). Therefore, our study defines the interplay between two chromatin-remodeling systems whose activity depends on the methylation state of H3K4 in the reorganization of chromatin and induction of gene expression. Histone methylation is a key histone modification relevant for gene regulation in normal and disease states. Our study illustrates the complexity of the regulation of chromatin remodeling by histone methylation for stress-induced gene expression unveiling a key role for H3K4 monomethylation. The inhibition of SWR1 by such methylation might have many implications to understand phenotypes associated with Set1 inactivation. This novel mechanism of regulation of chromatin remodeling found upon stress may have wider implications for chromatin remodeling and gene expression in additional scenarios and in other systems.
Expression of stress-responsive genes is controlled by two remodeling mechanisms, RSC and SWR-C, depending on the presence of monomethylated H3K4. Scheme summarizing a novel role for H3K4 monomethylation in dictating the specificity of chromatin remodeling at stress-responsive genes. When methylation is present (colored histones), RSC remodels chromatin to induce gene expression. In the absence of methylation (grey histones), Swr1 promotes chromatin remodeling and RSC become dispensable to induce stress-responsive gene expression.
We thank L. Subirana and A. Fernandez for technical support. We are grateful to Drs Christoph Schüller, Jerry Workman and Frank Holstege for helpful discussions.
FUNDING
M.N. was a recipient of an FIS fellowship. G.M.Z. was a recipient of an FPU fellowship from the Spanish Government. Spanish Ministry of Economy and Competitiveness [BFU2012–33503 and FEDER to F.P., BFU2011–26722 to E.N., BFU2013–48643-C3–1-P and FEDER to S.C.]; Catalan Government (2014 SGR 599) and the Andalusian Government [BIO-271; P12-BIO-1938 and FEDER to S.C.]. Fundación Botín, by Banco Santander through its Santander Universities Global Division to F.P. F.P. and E.N. are recipients of an ICREA Acadèmia (Generalitat de Catalunya). The authors declare no competing financial interest. Funding for open access charge: Spanish Ministry of Economy and Competitiveness [BFU2012-33503 and FEDER to F.P.].
Conflict of interest statement. None declared.




Comments