-
PDF
- Split View
-
Views
-
Cite
Cite
Jonathan R. Karr, Jayodita C. Sanghvi, Derek N. Macklin, Abhishek Arora, Markus W. Covert, WholeCellKB: model organism databases for comprehensive whole-cell models, Nucleic Acids Research, Volume 41, Issue D1, 1 January 2013, Pages D787–D792, https://doi.org/10.1093/nar/gks1108
Close - Share Icon Share
Abstract
Whole-cell models promise to greatly facilitate the analysis of complex biological behaviors. Whole-cell model development requires comprehensive model organism databases. WholeCellKB (http://wholecellkb.stanford.edu) is an open-source web-based software program for constructing model organism databases. WholeCellKB provides an extensive and fully customizable data model that fully describes individual species including the structure and function of each gene, protein, reaction and pathway. We used WholeCellKB to create WholeCellKB-MG, a comprehensive database of the Gram-positive bacterium Mycoplasma genitalium using over 900 sources. WholeCellKB-MG is extensively cross-referenced to existing resources including BioCyc, KEGG and UniProt. WholeCellKB-MG is freely accessible through a web-based user interface as well as through a RESTful web service.
INTRODUCTION
A primary challenge in computational biology is to predict how complex phenotypes such as growth and replication arise from networks of individual molecules. Whole-cell models promise to tackle this challenge by integrating heterogeneous molecular data into predictive computational models. This integration requires model organism databases which comprehensively provide readily computable molecular data.
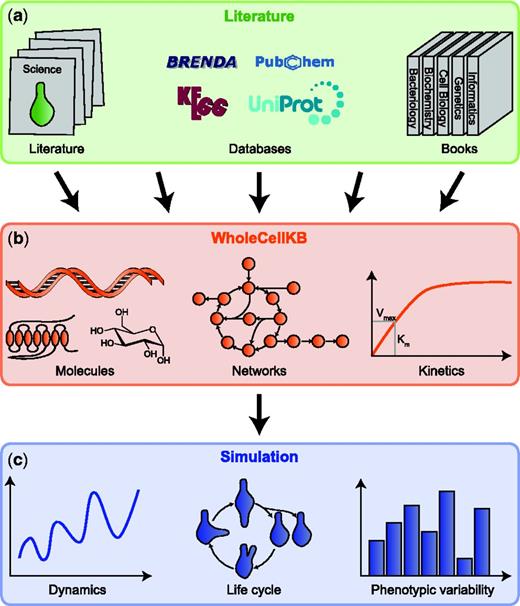
WholeCellKB is an open-source, web-based software program for developing comprehensive model organism databases for whole-cell models. As illustrated in Figure 1, WholeCellKB enables whole-cell modeling by organizing diverse molecular data from primary research articles, reviews, books and databases into a single database. The WholeCellKB data model supports detailed descriptions of individual species including their genes, operons, proteins, macromolecular complexes, molecular interactions, chemical reactions and pathways. Importantly, WholeCellKB also facilitates extensive source documentation. We used WholeCellKB to develop WholeCellKB-MG, an extensive database of the pathogenic Gram-positive bacterium Mycoplasma genitalium.
WholeCellKB-MG enables whole-cell modeling by integrating diverse data sources into a single database. (a) Currently, WholeCellKB-MG integrates >900 primary research articles, reviews, books and databases. (b) WholeCellKB-MG comprehensively represents all aspects of molecular physiology including metabolomics, genomics, transcriptomics and proteomics. (c) WholeCellKB-MG provides molecular data for whole-cell models.
Here, we describe WholeCellKB-MG’s content, curation, user interface and implementation. We also compare WholeCellKB-MG to existing resources, highlighting WholeCellKB-MG’s greater scope and granularity. Finally, we discuss our future plans for WholeCellKB.
CONTENT
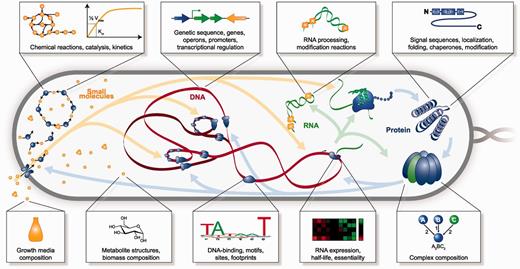
Our goal was to create a database comprehensive enough to enable a whole-cell model (1). As illustrated in Figure 2, WholeCellKB-MG broadly represents M. genitalium molecular biology including (i) its subcellular organization; (ii) its chromosome sequence; (iii) the location, length, direction and essentiality of each gene; (iv) the organization and promoter of each transcription unit; (v) the expression and degradation rate of each RNA transcript; (vi) the specific folding and maturation pathway of each RNA and protein species including the localization, N-terminal cleavage, signal sequence, prosthetic groups, disulfide bonds and chaperone interactions of each protein species; (vii) the subunit composition of each macromolecular complex; (viii) its genetic code; (ix) the binding sites and footprint of every DNA-binding protein; (x) the structure, charge and hydrophobicity of every metabolite; (xi) the stoichiometry, catalysis, coenzymes, energetics and kinetics of every chemical reaction; (xii) the regulatory role of each transcription factor; (xiii) its chemical composition and (xiv) the composition of its laboratory growth medium. Table 1 summarizes WholeCellKB-MG’s size and content.
WholeCellKB aims to comprehensively describe cell physiology including the structure and dynamics of every metabolite, gene, RNA transcript and protein. Boxes illustrate several molecular properties represented by WholeCellKB.
WholeCellKB-MG size
| Entry type . | Number . |
|---|---|
| Cellular state | 16 |
| Chromosome feature | 2305 |
| Compartment | 6 |
| Gene | 525 |
| Metabolite | 722 |
| Pathway | 17 |
| Process | 28 |
| Protein complex | 201 |
| Protein monomer | 482 |
| Reaction | 1857 |
| Transcription unit | 335 |
| Transcriptional regulatory interaction | 30 |
| Entry type . | Number . |
|---|---|
| Cellular state | 16 |
| Chromosome feature | 2305 |
| Compartment | 6 |
| Gene | 525 |
| Metabolite | 722 |
| Pathway | 17 |
| Process | 28 |
| Protein complex | 201 |
| Protein monomer | 482 |
| Reaction | 1857 |
| Transcription unit | 335 |
| Transcriptional regulatory interaction | 30 |
WholeCellKB-MG size
| Entry type . | Number . |
|---|---|
| Cellular state | 16 |
| Chromosome feature | 2305 |
| Compartment | 6 |
| Gene | 525 |
| Metabolite | 722 |
| Pathway | 17 |
| Process | 28 |
| Protein complex | 201 |
| Protein monomer | 482 |
| Reaction | 1857 |
| Transcription unit | 335 |
| Transcriptional regulatory interaction | 30 |
| Entry type . | Number . |
|---|---|
| Cellular state | 16 |
| Chromosome feature | 2305 |
| Compartment | 6 |
| Gene | 525 |
| Metabolite | 722 |
| Pathway | 17 |
| Process | 28 |
| Protein complex | 201 |
| Protein monomer | 482 |
| Reaction | 1857 |
| Transcription unit | 335 |
| Transcriptional regulatory interaction | 30 |
CURATION
We curated WholeCellKB-MG in five steps based on >900 primary research articles, reviews, books and databases. First, we curated the overall structure of M. genitalium including its size, shape, subcellular organization and chemical composition based on several experimental studies including Morowitz et al. (2). We also assembled the chemical composition of Mycoplasma laboratory growth medium based on analyses reported by Solabia (3).
Second, we curated the structure of the M. genitalium chromosome including its sequence, the location, length and direction of each gene and its transcription unit organization based on the Comprehensive Microbial Resource (CMR) annotation (4) and a recent study by Güell et al. (5). We reconstructed the location of each promoter and the expression, degradation rate and essentiality of each gene product from four recent studies (6–9). We catalogued DNA-binding sites and transcriptional regulatory interactions from several sources including DBTBS (10).
Third, we assembled the structure of each RNA and protein gene product. We compiled the post-transcriptional processing and modification of each RNA transcript from several sources including Peil (11). We reconstructed the signal sequence, localization, chaperone-mediated folding, post-translational modification, disulfide bonds, subunit composition and DNA footprint of each protein and macromolecular complex from a large number of primary research articles, computational models and databases. We assembled the chemical regulation of each gene product from several sources including DrugBank (12). We used ExPASy ProtParam (13) to calculate the pI, extinction coefficient, half-life, instability index, aliphatic index and grand average of hydropathy of every protein species.
Fourth, we curated the specific chemical reactions catalyzed by each gene product starting from the CMR (4), GenBank (14), KEGG (15) and UniProt (16) genome annotations and the reconstructed RNA and protein maturation pathways. To maximize the scope of the database and to fill gaps in the genome annotation, we expanded each gene product’s annotation based on primary research articles we identified by searching PubMed (17) and Google Scholar (http://scholar.google.com). We consulted BioCyc (18), KEGG (15), two flux-balance analysis (FBA) models of bacterial metabolism (19,20) and hundreds of additional primary research articles to curate the stoichiometry of each chemical reaction. We assembled the thermodynamics and kinetics of each chemical reaction from several databases including BRENDA (21), SABIO-RK (22) and UniProt (16) and a FBA model (20).
Finally, we compiled the M. genitalium metabolome. We included all metabolites involved in the reconstructed reactions, biomass or growth medium. We curated the empirical formula, structure, charge and intracellular concentration of each metabolite from several databases including BioCyc (18), CyberCell (23) and PubChem (24) and a comprehensive mass-spectrometry study (25). We used ChemAxon Marvin (http://www.chemaxon.com/products/marvin) to calculate the molecular weight, van der Waals volume, pI, logd and logp of each metabolite.
In order to create a comprehensive description of M. genitalium physiology, we based WholeCellKB-MG on studies of closely related organisms where studies of M. genitalium were unavailable. In cases where multiple observations were available, we based the reconstruction on the most closely related organism. We used bi-directional best BLAST (26) to identify homologous genes. To provide model transparency, we tracked the species, experimental conditions and citation of each piece of evidence.
COMPARISON TO EXISTING RESOURCES
WholeCellKB represents the specific molecular interactions of individual species similar to previous databases such as BioCyc (18,27) and BiGG (28). In particular, WholeCellKB’s data model, user interface and species-specific content were heavily inspired by BioCyc.
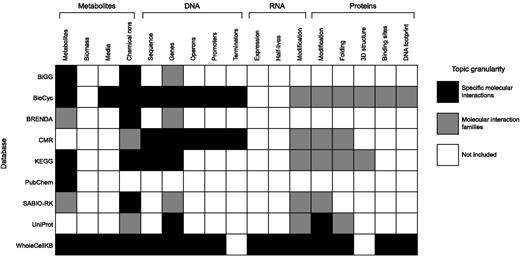
Importantly, WholeCellKB-MG also has several major differences from existing resources. First, WholeCellKB-MG more broadly represents cell physiology. WholeCellKB-MG represents the molecular details of 28 cellular processes including well-studied processes such as metabolism as well as less well-understood processes such as DNA damage and repair and RNA and protein degradation. The online documentation at http://wholecellkb.stanford.edu/about provides further information about the WholeCellKB-MG data model and how WholeCellKB-MG represents each cellular process. Figure 3 compares WholeCellKB-MG’s content to that of several existing databases.
Detailed comparison of the content of WholeCellKB-MG and several existing biological databases. In addition to containing detailed descriptions of genetics, metabolism and transcriptional regulation comparable to existing resources such as BiGG (28), BioCyc (18) and CMR (4), WholeCellKB-MG has detailed representations of RNA degradation, RNA and protein maturation and protein translocation. Black boxes indicate physiology represented with fine granularity including the specific molecules involved in each specific interaction (e.g. specific metabolites involved in each metabolic reaction). Gray boxes indicate coarsely represented physiology, for example lumping families of similar reactions such as RNA methylation into a single database entry rather than representing the specific RNA bases involved in each individual reaction. White boxes indicate unrepresented physiology.
Second, whole-cell modeling requires model organism databases which explicitly define the participants of each molecular interaction and chemical reaction. WholeCellKB-MG addresses this need by representing the specific molecules involved in every molecular interaction and by requiring structures for each molecule. For example, WholeCellKB-MG represents the specific RNA bases involved in every RNA methylation reaction, whereas existing resources lump RNA methylation interactions into a single generic reaction. WholeCellKB-MG represents every major cellular process including RNA processing and protein processing, modification and translocation with similarly fine molecular resolution.
Third, where available WholeCellKB-MG contains not only structural but also quantitative functional descriptions of each molecule and molecular interaction. For example, WholeCellKB-MG contains chemical reaction rate laws and kinetic parameters, RNA transcript expressions and half-lives, and cellular and growth medium chemical compositions. In total, WholeCellKB-MG represents 1836 heterogeneous model parameters. Table 2 summarizes how WholeCellKB represents these heterogeneous parameters using several types of database entries.
WholeCellKB-MG parameters
| Type . | Number . |
|---|---|
| Cell composition | 73 |
| Media composition | 83 |
| Reaction Keq | 225 |
| Reaction Km | 483 |
| Reaction Vmax | 434 |
| RNA expression | 525 |
| RNA half-life | 525 |
| Stimulus values\ | 10 |
| Transcriptional regulation | 32 |
| Activity | 30 |
| Affinity | 2 |
| Other | 154 |
| Type . | Number . |
|---|---|
| Cell composition | 73 |
| Media composition | 83 |
| Reaction Keq | 225 |
| Reaction Km | 483 |
| Reaction Vmax | 434 |
| RNA expression | 525 |
| RNA half-life | 525 |
| Stimulus values\ | 10 |
| Transcriptional regulation | 32 |
| Activity | 30 |
| Affinity | 2 |
| Other | 154 |
WholeCellKB-MG parameters
| Type . | Number . |
|---|---|
| Cell composition | 73 |
| Media composition | 83 |
| Reaction Keq | 225 |
| Reaction Km | 483 |
| Reaction Vmax | 434 |
| RNA expression | 525 |
| RNA half-life | 525 |
| Stimulus values\ | 10 |
| Transcriptional regulation | 32 |
| Activity | 30 |
| Affinity | 2 |
| Other | 154 |
| Type . | Number . |
|---|---|
| Cell composition | 73 |
| Media composition | 83 |
| Reaction Keq | 225 |
| Reaction Km | 483 |
| Reaction Vmax | 434 |
| RNA expression | 525 |
| RNA half-life | 525 |
| Stimulus values\ | 10 |
| Transcriptional regulation | 32 |
| Activity | 30 |
| Affinity | 2 |
| Other | 154 |
DATA INPUT
WholeCellKB provides administrators with two editing interfaces: (i) a web form to edit single entries and (ii) an Excel-based interface to simultaneously edit multiple entries. We believe that these two interfaces enable collaborative model organism database development.
In the beginning of our M. genitalium curation efforts, we primarily used the batch interface to quickly import large amounts of data from other genome annotations. We continued to use the batch interface throughout the project to import high-throughput molecular data. Later in our M. genitalium curation efforts, we primarily used the form interface to refine our annotation based on specific biochemical studies. Overall, we found that WholeCellKB improved the quality of our annotation and in particular encouraged us to thoroughly annotate the original source of each datum.
Data submitted to WholeCellKB was extensively validated to ensure consistency and correctness. For example, WholeCellKB checked that each chemical formula was valid, that each reaction was mass-balanced and that every molecule and kinetic parameter was defined in each reaction rate law. WholeCellKB provided hints on how to correct invalid data such as the atom imbalance of invalid reactions.
DATA ACCESS
WholeCellKB-MG is freely accessible through a simple and intuitive web-based interface at http://wholecellkb.stanford.edu. This web-based interface allows users to quickly browse, search and export the database. It also allows administrators to add, edit and delete entries. Importantly, the interface is extensively commented and hyperlinked, allowing users to easily find the primary source of each datum.
WholeCellKB-MG is also accessible through a RESTful interface. This interface provides the content of every HTML page in JSON and XML formats. We are currently using this interface to develop software for visualizing whole-cell simulations.
DEVELOPER API
WholeCellKB was designed to enable modelers to develop model organism databases for whole-cell models, including designing custom data models and user interfaces. WholeCellKB provides a framework for viewing, searching, exporting and editing database entries which developers can combine with custom data models and HTML templates. This allows developers to build custom model organism databases with minimal effort and without any knowledge of database design. Furthermore, because WholeCellKB is open source and implemented with Python, modelers can easily display scientific calculations alongside curated data in the user interface. The online documentation provides further instructions on how to customize WholeCellKB.
IMPLEMENTATION
WholeCellKB was implemented in Python using the Django (http://www.djangoproject.com) web framework and stored using the relational database MySQL (http://www.mysql.com). Full-text search was implemented using Haystack (http://haystacksearch.org) and Xapian (http://xapian.org). Excel, JSON and XML export were implemented using OpenPyXL (http://bitbucket.org/ericgazoni/openpyxl), simplejson (http://pypi.python.org/pypi/simplejson) and xml.dom (http://docs.python.org/ library/xml.dom.html). WholeCellKB runs on the Apache (http://www.apache.org) web server using the mod_wsgi (http://code.google.com/p/modwsgi) module. All of the software used to implement WholeCellKB is available open source.
SUMMARY AND FUTURE DIRECTIONS
WholeCellKB-MG is an extensive database of M. genitalium designed to facilitate whole-cell modeling. Currently, we are continuing to curate the database as well as starting to create equally comprehensive databases of other model microorganisms. Beyond facilitating realistic whole-cell models, we believe that these databases are useful platforms for experimental and computational biologists.
We created WholeCellKB-MG using WholeCellKB, an open-source, web-based software program which enables modelers to quickly develop model organism databases for whole-cell modeling.
Beyond continuing to curate model organisms, we also plan to continue to strengthen the WholeCellKB software. We plan to add additional tools for importing databases curated with other tools such as PathwayTools (27), storing the detailed history of each database entry and comparing model organism databases as well as expanding the search functionality of the RESTful API. As the whole-cell modeling community grows, in the future we also plan to enable open-editing similar to Wikipedia. Finally, we are currently using WholeCellKB’s RESTful API to develop tools for visualizing whole-cell simulations.
We hope that other researchers will use WholeCellKB to develop model organism databases and whole-cell models. We believe that WholeCellKB will not only speed up database curation and whole-cell model development but also encourage best annotation practices. Ultimately, we hope that WholeCellKB in combination with whole-cell models will accelerate biological discovery and bioengineering.
FUNDING
NIH Director’s Pioneer Award [5DP1LM01150-05] and a Hellman Faculty Scholarship (to M.W.C.); NDSEG, NSF and Stanford Graduate Fellowships (to J.R.K.); NSF and Bio-X Graduate Student Fellowships (to J.C.S.) and a Stanford Graduate Fellowship (to D.N.M.). Funding for open access charge: NIH Director’s Pioneer Award [5DP1LM01150-05].
Conflict of interest statement. None declared.
ACKNOWLEDGEMENTS
We thank Elsa Birch, Nick Ruggero and Ruby Lee for enlightening discussions on database design, curation, modeling and visualization.




Comments