-
PDF
- Split View
-
Views
-
Cite
Cite
Chenli Liu, Claudia Danilowicz, Nancy Kleckner, Mara Prentiss, Single molecule identification of homology-dependent interactions between long ssRNA and dsDNA, Nucleic Acids Research, Volume 45, Issue 2, January 2017, Pages 894–901, https://doi.org/10.1093/nar/gkw758
Close - Share Icon Share
Abstract
Long non-coding RNAs (lncRNAs) are prominently associated with chromosomes in an ever-increasing diversity of roles. To provide further insight into the potential nature of these associations, we have explored, for the first time, the interaction of long single-stranded (ss) RNAs with cognate homologous double-stranded (ds) DNA in vitro. Using magnetic tweezers, we measured the effects of ssRNA on force extension curves for dsDNA. We observe that the presence of ssRNA impedes the extension of dsDNA, specifically at low forces, dependent on homology between the RNA and DNA species, and dependent on ssRNA lengths (≥1 kb). The observed effect also depends on the concentration of ssRNA and is abolished by overstretching of the dsDNA. These findings show that significant homologous contacts can occur between long ssRNA and dsDNA in the absence of protein and that these contacts alter the mechanical properties of the dsDNA. We propose that long ssRNA interacts paranemically with long dsDNA via periodic short homologous interactions, e.g. mediated by RNA/DNA triplex-formation, and that dsDNA extension is impeded by formation of RNA secondary structure in the intervening unbound regions. Analogous interactions in vivo would permit lncRNAs to mediate the juxtaposition of two or more DNA regions on the same or different chromosomes.
INTRODUCTION
Five decades ago, Bonner et al. first reported that RNA associated with chromatin (1). Since then, an increasing body of studies has showed that interactions between RNA and chromosomes play important roles in vivo, e.g. for DNA repair, transcriptional gene regulation, post-transcriptional processing, chromatin modification, etc. (2–5).
For the mammalian transcriptome, long noncoding RNAs (lncRNAs) comprise a main subgroup (6). It has been shown that lncRNAs form scaffolds with chromatin to mediate epigenetic regulation and to form higher-order chromatin structure and functional nuclear sub-compartments (5). Interestingly, also, in fission yeasts, an interchromosomal interaction mediated by sme2 RNA plays an important role in pairing of homologous chromosomes (2). It has been suggested that the scaffolds formed by lncRNAs are mobile, thus permitting their interaction with multiple gene loci; alternatively or in addition, these RNAs are suggested to act as organizing centers, gathering multiple gene loci into higher-order chromatin structure (5).
The mechanisms by which lncRNAs interact with chromosomes are only beginning to emerge. In vitro studies using oligonucleotide substrates have shown that single-stranded (ss) RNA is capable of specifically binding homologous double-stranded (ds) dsDNA through formation of a triple helix, where the ssRNA accommodates itself in the major groove of dsDNA (7). These RNA-DNA triple-helices were suggested to be involved in the biological roles of non-coding RNA (8,9). Following on this possibility, local single-site triplex formation has recently been implicated in the interactions of ncRNAs with their cognate DNA target regions, with inferences drawn by a combination of oligonucleotide-based assays and correlated in vivo effects (8,10,11).
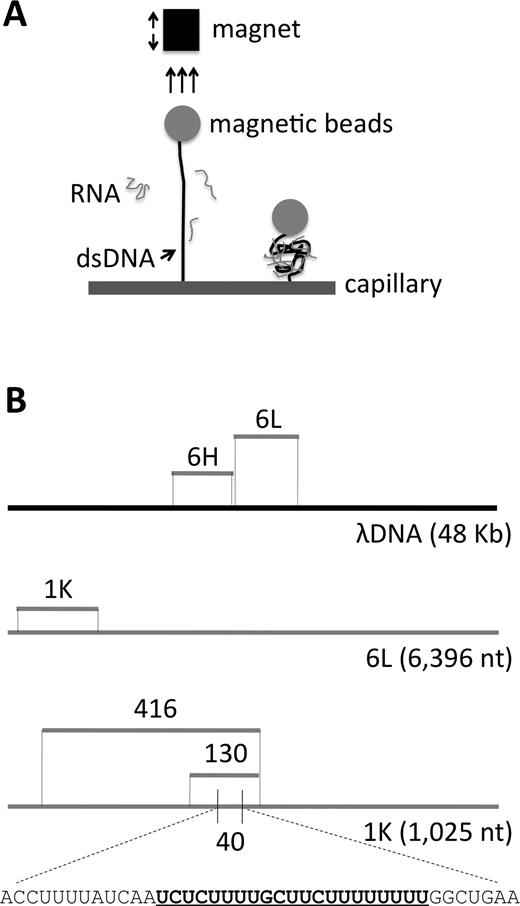
The above studies leave unaddressed the possibility that long ssRNA might interact with an homologous dsDNA in more complex ways, either in vitro or in vivo. Here, we begin to explore this long molecule regime in vitro. Using our previously-described magnetic tweezers apparatus (12,13), we have defined the force extension curve of dsDNA in the absence, and in the presence, of various types of ssRNAs, at physiological (neutral) pH, with the goal of detecting changes in the dsDNA behaviour that might imply the presence of an interaction (Figure 1A).
Schematic diagram of the experimental design. (A) Schematic representation of the assay for measuring the extension of dsDNA due to the binding of RNA. (B) Sequences to which analyzed RNAs are homologous. 6L = 6,396-nt low G+C content RNA and 6H = 6,107-nt high G+C content RNA; positions shown relative to full length λ DNA template (black line). 1K = a 1025-nt subsegment of 6L. 416, 130 and 40 are subsegments of 1K. Gray lines denote ssRNAs. A predicted nucleic acid triple helices target site in the 40nt RNA is bold and underlined within the total sequence.
MATERIALS AND METHODS
Apparatus
The magnetic tweezers apparatus was set up as previously described (13). The magnets were held in a lateral position with respect to a microchannel containing the sample on a three-axis translation stage to exert a force perpendicular to the glass surface to which the dsDNA was bound. The magnitude of the force applied on the beads was determined by the distance between the magnet and the glass surface. The calibration of the relationship between the force on a magnetic bead and the position of the magnet was originally established using measurements of the Stokes drag on individual magnetic beads in glycerol. This calibration was checked using a Gauss meter to measure the magnetic field as a function of distance from the magnet and using the known magnetization of the magnetic beads to calculate the force. The beads have some variation in magnetization, which means that different beads experience slightly different forces at a given magnet position. The variation in the force is found to be ±5% for these beads. As a result, the distance between the bead and the capillary, as a function of force, varies from bead to bead. For the sake of clarity, we normalized the force extension to 14 μm at 10 pN for each bead. This normalization does not affect the shapes of force extension curves. Occasionally, more than one dsDNA molecule will bind to a single bead. Such cases can be detected by subjecting the bead to high forces that are sufficient for a single dsDNA/bead ensemble to cause the dsDNA to undergo the structural transition of ‘overstretching’, which is detected as a large increase in extension at forces above 65 pN. Thus, upon completion of experimental assessment, every bead/dsDNA ensemble was assayed for the overstretching response, as well as for the particular behavior of interest. Ensembles that fail to exhibit the diagnostic overstretching response were not considered. For observation of dsDNA overstretching, a small magnet with higher magnetization was manually moved slowly towards the glass surface. The overstretching response is indicated by a discrete increase in DNA length, as described previously (14). All experiments were carried out at room temperature (∼25°C).
Sample preparation
Full-length λ dsDNA (NEB; 48 502 bp) was prepared as previously described (14). Briefly, λ dsDNA was hybridized and ligated to an oligonucleotide, complementary to the ssDNA tail at one of its ends, and containing a Digoxigenin-labeled oligonucleotide and to the ssDNA tail with a biotinylated oligonucleotide at the other end.
Total RNA was extracted from exponentially growing culture of Saccharomyces cerevisiae strain YS1267 using TRIzol (Invitrogen) and following the manufacturer's instructions. Other RNAs (Figure 1) were transcribed in vitro using T7 Quick High Yield RNA Synthesis Kit (NEB) and DNA templates prepared by amplification with Crimson LongAmp® Taq DNA polymerase (NEB). For short transcripts (<0.3 kb), incubation of amplification reactions was extended to 24 h. Long RNAs were purified by phenol-chloroform extraction and ethanol precipitation, and dissolved in DEPC-treated water. Short RNAs (130 and 416 nt) were purified by gel electrophoresis on a Criterion 10% polyacrylamide TBE–urea gel (Bio-Rad), precipitated with ethanol and redissolved in DEPC-treated water. The shortest RNA (40 nt) and other oligonucleotides used in this study were purchased from Integrated DNA Technologies. Oligonucleotide sequences are listed in Supplementary Table S1.
Interaction reaction
Interaction experiments were carried out in a square micro-cell (Vitrotubes; 0.8 mm) containing a round inner capillary (Vitrotubes; 0.55 mm in diameter) closed at its ends. The inner capillary was modified by adsorption of 1 mg/ml streptavidin in PBS (phosphate-buffered saline) pH 7.4 overnight. The reaction mixture containing dsDNA, ssRNA, RNasin® Ribonuclease Inhibitor (Promega) and superparamagnetic beads (Dynabeads® M-280 Streptavidin) was prepared and pre-incubated for 5 min at room temperature (∼25°C). The concentration of λ dsDNA was 1 μg/ml in all experiments. RNA concentrations were varied over a 105-fold range, e.g. 6L RNA analysis begins with 0.06 μg/ml, which represents a 1:1 molar ratio of RNA to DNA.
The reaction mixture was then introduced into the micro-cell. After an initial incubation of 5 min, the DNA molecules became tethered between the glass capillary surface and the streptavidin-coated beads. The micro-cell was then placed in the magnetic tweezers apparatus. Force was increased at 1-s time intervals in force steps of ΔF = 0.04 pN for forces less than 0.7 pN, ΔF = 0.06 pN for forces between 0.7 and 1.5 pN, and ΔF = 0.2 pN for higher forces. The position of each bead was followed in real time using an inverted microscope and a digital camera. Each bead was tracked by bead tracking software and recorded using Matlab as described previously (13). After observation was complete, the integrity of the RNA transcripts in the micro-well was tested by denaturing agarose gel electrophoresis. No significant decrease in the concentration or average RNA molecular weight was observed in any of the reported studies, implying absence of significant degradation.
Statistical analysis
The data and the variation among the data were represented using box and whisker plots. In brief, the data are arrayed in order; the median value is identified and shown as the horizontal line within the rectangular box; the medians are then taken for the sets of data above and below the median, and these medians (which also define the third and fourth quartiles of the data, respectively) define the top and bottom of the rectangular box; the vertical lines extending from the top and bottom of the rectangular box are set at 1.5× interquartile range above the third quartile, and 1.5× interquartile range below the first quartile. For these plots, statistical significance was analyzed by unpaired Student's t-test, with P-values <0.05 considered statistically significant.
RESULTS
Long ssRNA changes the mechanical properties of dsDNA
The response of dsDNA to progressively increasing force (‘force–extension analysis’) has been precisely described in many previous studies. Here, we investigate the effects of pre-incubating an ssRNA with the dsDNA prior to defining its force extension response. For such analysis (13; Figure 1A), the two ends of full-length λ dsDNA are differentially labeled, one with biotin and the other with digoxigenin. The digoxigenin-labeled end can attach specifically to an anti-digoxigenin coated capillary, and the biotin-labeled DNA can attach specifically to a streptavidin coated magnetic bead. Force is imposed to extend the λ dsDNA by adjusting the position of a magnet. For experimental analyses, an RNA species of interest is present in the capillary chamber prior to the application of force; control experiments for λ dsDNA in the absence of RNA were also performed. Imposition of force moves the bead away from the surface of the capillary (Figure 1A). The bead/surface distance measured by using an inverted microscope and a digital camera (see Methods)(12). All ssRNAs used in this study were transcribed in vitro from certain regions of the λ genome (Figure 1B).
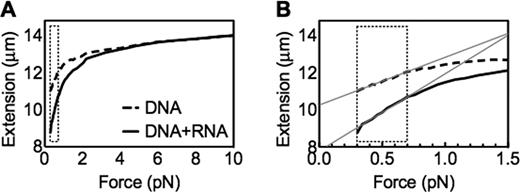
In our apparatus, forces between 0.4 and 0.7 pN can extend λ dsDNA to between 12 and 13 μm (13). Here, we progressively increased the imposed force starting from 0.3 pN. At this lowest force, the distance between the bead and the capillary is approximately 11 μm for λ dsDNA alone (Figure 2A, dashed line). In contrast, in the presence of a 6,396 nt ssRNA homologous to an internal region of the λ dsDNA (RNA ‘6L’; Figure 1B), the distance is ∼3 μm shorter (Figure 2A, solid line). This result suggests that there has been an interaction between the long ssRNA and the dsDNA which somehow impedes the typical dsDNA force-mediated extension. At higher forces, the force–extension curve obtained with RNA/dsDNA co-incubation approaches that of λ dsDNA alone, with the two curves converging at ∼6 pN. Thus, the effect of 6L RNA on homologous dsDNA extension is only evident at low forces, consistent with disruption of the previously-established interaction at forces above ∼6 pN.
Long ssRNA changes the mechanical property of dsDNA. (A) Representative single force extension curves of λ dsDNA in the presence or absence of long ssRNA 6L (DNA:RNA mol ratio = 1:1000); (B) a zoom-in curve of (A). The dashed brackets indicate the low force region (0.3–0.7 pN). The grey lines indicate the linear trend lines at this region, by which the slopes in Figures 3 and 4 are calculated.
At the end of a series of measurements for one bead/dsDNA ensemble, we performed an additional test to determine whether the bead was attached to the capillary by one single dsDNA molecule. To perform this test, we exploited the overstretching transition that is characteristic of dsDNA. Overstretching is a force induced increase in length that extends the dsDNA to ∼1.7× the B-form length. The transition occurs at an applied force of ∼65 pN, and the width of the transition is ∼2 pN. If one single dsDNA binds the bead to the surface, then applying a force of ∼65 pN to the bead will increase the extension of the dsDNA by a factor of ∼1.7. In contrast, if the bead is bound to the surface by two dsDNA, then the applied force will be shared by both dsDNA molecules, then a force of ∼130 pN would be required to overstretch both dsDNA molecules. Thus, observation of an abrupt length increase at an applied force of ∼65 pN provides confirmation that the dsDNA is bound to the surface by a single dsDNA molecule.
The low force experiments were done with a stack of permanent magnets with a low magnetic field gradient. That low gradient results in a low applied force, but the low gradient improves the accuracy of the force values at forces <10 pN. The ∼65 PN force required to overstretch dsDNA could not be reached using the magnet stack we used for the low force measurements, so overstretching was achieved by adding a small magnetic tip to the stack of permanent magnets after the low force measurements were completed. The magnetic tip increases the field gradients, allowing us to apply a force in excess of 70 pN. In the presence of the magnetic tip, the applied force is poorly calibrated; however, the force due to the tip cannot exceed 100 pN. After the magnetic tip was attached to the stack of magnets, the magnets and the attached tip were moved toward the capillary. That motion toward the capillary steadily increases the magnetic force on the bead. The approach toward the capillary continued until overstretching was observed as a sudden change in extension of the tethered molecule to about 1.7 times its contour length. This significant change in extension is easily observed; if more than one dsDNA molecule is tethered between the magnetic bead and the capillary surface, the force applied with the magnetic tip is insufficient to overstretch the dsDNA. Thus, any bead that did overstretch was assumed to be tethered to the surface by a single dsDNA, whereas any bead that did not overstretch was assumed to be tethered to the surface by more than one dDNA. Any data taken with a bead tethered by more than one dsDNA was rejected, so all of the data presented in this paper corresponds to experiments where the magnetic bead was attached to the surface by one single dsDNA molecule.
We note that interactions between ssRNA 6L and λ dsDNA could cause a decrease in the slope of the force–extension curve by changing the stiffness of the dsDNA per se and/or by forming secondary structures in which different dsDNA regions are linked. Changes in extension due to the formation of secondary structures have been observed for single-stranded DNA (ssDNA). In particular, at low forces, hairpin formation results in decreases in extension; however, when the applied force is sufficient to melt all of the hairpins, the extension versus force curve is no longer influenced by the presence of secondary structure. For ssDNA, the force required to melt hairpins is ∼9 pN (15,16). In the present case, we suggest that the observed effect could result from local interactions of the ssRNA at multiple positions along the dsDNA, in combination with ssRNA hairpins (Discussion).
We also note that, given these possibilities, we cannot characterize the observed extension vs. force curves in terms of ‘persistence length’. Such characterization is possible for force versus extension curves of an individual polymer that is not interacting with itself (a ‘non-interacting polymer’). The force versus extension curves for non-interacting polymers are well characterized by entropic models based on non-interacting units with length of the order of the persistence length. In the present case, however, the dsDNA is, effectively, a self-interacting polymer, and such polymers have force versus extension curves that contain additional free energy terms due to interactions between different parts of the polymer (15). Thus, we cannot characterize the extension versus force curves in terms of ‘persistence length’ as defined by a model based on non-interacting polymers. For this reason, in what follows, we simply report the experimentally observed changes in the extension versus force curve without fitting those curves to any specific model (e.g. a model based on non-interacting polymers).
The effect of homologous ssRNA on extension of dsDNA at low force is RNA concentration dependent
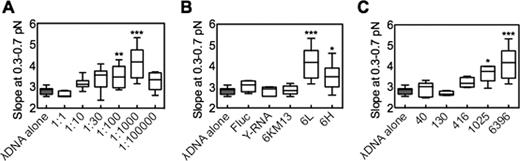
We performed force–extension analyses analogous to those described above, with 1 μg/ml dsDNA per reaction mixture, but with varying amounts of RNA 6L in molar ratios ranging from 1:1 to 1:100 000, measuring the slope (x/F) at 0.3–0.7 pN of λ dsDNA as shown in Figure 2B. As can be seen in Figure 3A, at a 1:1 DNA/RNA ratio, no effect of the RNA is detectable. At ratios of 1:10–1:10,000, the slope of the force–extension curve progressively increases. A strong decrease then occurs at the highest ratio (1:100 000). The basis for this decrease is unclear. One possible explanation is that the high concentration of ssRNA leads to condensate formation, which prevents RNA binding to dsDNA. However, there is another, more interesting, possibility. We suggest that the effect observed at lower DNA/RNA ratios is due to the interaction of one ssRNA molecule with distant dsDNA binding sites that are brought together by the folding of the ssRNA; moreover, it is likely that the number of available binding sites is limited (Discussion). In this situation, at high RNA concentration, there will be an increased probability that the available sites will be bound independently by different ssRNAs, thus precluding tethering of disparate dsDNA sites. In contrast, at low ssRNA concentrations binding of RNA to DNA may be sufficiently rare that a single ssRNA molecule that binds to one region has enough time to explore binding sites on the dsDNA and find a second binding position before another RNA binds to that region from the solution.
The effect of long ssRNA on dsDNA extension at low force (e.g. Figure 2B) is dependent on concentration, homology, and length. Box-and-whisker plots (see Materials and Methods) show the slopes of the low force–extension curves (Figure 2B) of the indicated samples. (A) DNA:RNA molar ratio was varied from 1:1 to 1:105. Statistical analysis (asterisks) compared 1:1 versus all other ratios. (B) The effects of non-homologous ssRNA controls. Fluc is a 1766-nt control transcript provided in the T7 Quick High Yield RNA Synthesis Kit, showing 43% identity with the 37 441–39 524 region of the genome of λ dsDNA. Y-RNA is the total RNA of S. cerevisiae strain. For 6KM13, bacteriophage M13mp18 circular ssDNA ∼7200 nt was amplified as the template for in vitro transcription. The resulting ssRNA is 6001 nt and presents 44% identity with the genome of λ dsDNA (region 22 893–31 255). 6L and 6H are ∼6k nt species homologous to adjacent regions of λ dsDNA (Figure 1B). DNA:RNA molar ratio = 1:1000 in all samples; statistical analysis was performed for the ratio of λ dsDNA alone versus all other ratios. (C) The effects of various lengths of ssRNA (described in Figure 1B), DNA:RNA molar ratio = 1:1000. (a, b, c) n = 10, *P < 0.01, **P < 0.001, ***P < 0.0001.
Long ssRNA impedes extension of dsDNA at low force only when the two species are homologous
The 6L ssRNA analyzed above is homologous to a sub-region of λ dsDNA. We next asked whether the observed effect is dependent on that homology. Several RNA transcripts were chosen for parallel analysis as non-homologous controls (Figure 3B). Fluc is a 1766-nt control transcript provided in the T7 Quick High Yield RNA Synthesis Kit, showing 43% identity with the 37 441–39 524 region of the genome of λ dsDNA. Y-RNA is the total RNA of S. cerevisiae strain. For 6KM13, bacteriophage M13mp18 circular ssDNA ∼7200 nt was amplified as the template for in vitro transcription. The resulting ssRNA is 6001 nt and presents 44% identity with the genome of λ dsDNA (region 22 893–31 255). None of these non-homologous RNAs changes the slope of the dsDNA force extension curve (Figure 3B). In contrast, a significant interaction was detected with a second homologous long ssRNA, 6H, which maps next to 6L along the λ dsDNA genome (Figures 1B and 3B). These results suggest the effect of the long ssRNAs on extension of dsDNA is homology-dependent.
The effect of homologous ssRNA on extension of dsDNA at low force increases with increasing RNA length
To determine if short ssRNAs have the same effect as long ones, we measured the slope of the low force–extension relationship for λ dsDNA coincubated with ssRNAs of different lengths (40, 130, 416, 1025 and 6396 nt; Figure 1B). We find that the slope significantly increases as the length of ssRNA is longer than 1,000 nt (Figure 3C). Thus, the ability of an ssRNA to alter dsDNA force extension curves is specifically a function of ssRNA length.
Overstretching eliminates the effect of long ssRNA on dsDNA
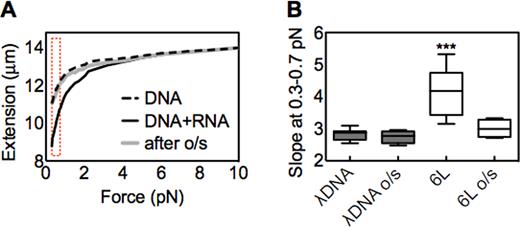
The above findings suggest that homology-based interaction of long ssRNA with dsDNA creates an impediment to force-mediated extension of the DNA. As discussed above, when dsDNA is subjected to pulling forces above ∼65 pN, its structure changes into an ‘overstretched’ conformation wherein its contour length is suddenly increased up to 1.7 times the canonical B-form dsDNA length. We were interested to know whether RNA/dsDNA interactions would, or would not, survive such a change in conformation. As discussed above, the maximum force that can be obtained using the stack of magnets is less than the force required to overstretch dsDNA. Thus, after studying the low force extension of the dsDNA, we added a magnetic tip to the magnetic stack. With the tip attached to the stack, we applied the ∼65 pN force required to overstretch the dsDNA. After overstretching, we withdrew the stack of magnets and the magnetic tip from the capillary until the applied force was <1 pN. We then removed the magnetic tip from the magnetic stack. With the tip removed, we took a second set of extension versus force curves using the stack of magnets to exert low forces on the magnetic bead. Several bead/dsDNA ensembles were probed in this way, and a typical result is shown in Figure 4.
Overstretching (o/s) abolishes the effect of long ssRNA. (A) Representative force extension curves in the presence or absence of ssRNA 6L (DNA:RNA mol ratio = 1:1000). One bead/dsDNA ensemble was initially followed and extended till about 10 pN (black solid curve). Subsequently a small tip was placed on the stack of magnets to achieve forces till about 70 pN (Materials and Methods; text). This bead/dsDNA exhibited a sudden change in extension that corresponds to the overstretching transition (not shown). The magnet was then slowly moved away from the glass surface to allow the molecule to recoil, and the tip was removed from the stack of magnets. Finally a second curve was measured with the stack of magnets up to a force of about 10 pN (gray curve). A typical force–extension curve for λ dsDNA in the absence of RNA is shown for comparison (dashed curve). The dashed bracket indicates the low force region (0.3–0.7 pN). (B) The box-and-whisker plot (Materials and Methods) displays the slope changes at low force region (0.3–0.7 pN) before or after overstretching (o/s). The statistical analysis was performed for λ dsDNA before overstretching versus all other samples. n = 10, ***P < 0.0001.
Beads that exhibited the signature diagnostic of RNA/DNA interaction when they first experienced low force did not retain that behavior after being subjected to overstretching. Instead, the force extension curve has the slope characteristic of dsDNA alone (Figure 4A), consequently the slope of overstretched samples significantly drops (Figure 4B). This result implies that the overstretching disrupts an existing homologous long ssRNA/dsDNA interaction, thus further confirming the presence of such an interaction. We also note that a detectable ssRNA/dsDNA interaction does not re-form after overstretching. This effect does not result from ssRNA degradation (Methods). Also, allowing increased time for recovery after overstretching did not restore a detectable interaction. We tested various waiting time periods (5, 10, 30 and 60 min) at zero force after the overstretching event and before another force increase; however, in no case did we observed the decrease in length at low force that we believe is characteristic of the binding of one ssRNA to multiple distant regions in the dsDNA. Its basis remains to be determined. However, this may result from different RNA molecules binding to the distant DNA binding sites as mentioned above.
DISCUSSION
The results presented demonstrate that long ssRNA and dsDNA can directly interact in a homology- and concentration-dependent manner without requiring the presence of proteins. This interaction is detected because it alters the typical extension of dsDNA at low forces. Interestingly, the detected interaction is specifically dependent on ssRNA length - it is prominent for RNAs of ∼1 kb and ∼6 kb and absent or greatly reduced for shorter RNAs.
A possible general explanation for these observations is as follows (Figure 5A). The ssRNA would be bound intermittently to the dsDNA along the homologous region. In between these interspersed contact points, the RNA would form secondary structures, creating bends in the dsDNA. The need to dissociate this secondary structure would explain why higher forces are required to get a particular dsDNA extension when homologous ssRNA is present than for dsDNA alone. Also, overstretching of the dsDNA in the ssRNA/dsDNA complex would dissociate the bound ssRNA molecule, allowing dsDNA to exhibit its typical force–extension behavior, as is observed.
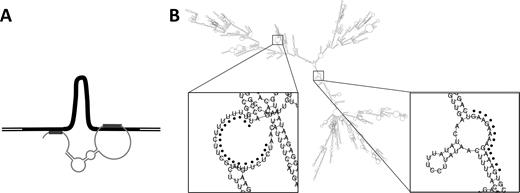
Possible inter-molecular interaction between long ssRNA and dsDNA. (A) Schematic diagram showing dsDNA with filled black segment indicating the region of homology with the ssRNA. The thin grey curve indicates the long ssRNA, which contacts the dsDNA at two positions (indicated by thick grey lines) and is folded into a secondary structure, thereby linking disparate segments of the dsDNA with concomitant looping of the intervening region. (B) A predicted secondary structure of 6L RNA via RNAfold webserver (http://rna.tbi.univie.ac.at/cgi-bin/RNAfold.cgi). Two enlarged regions containing predicted triplex target sites are indicated by black dots. A total of 11 such sites are predicted for this species (see text).
A further issue raised by these findings is the local nature of the constraining ssRNA/dsDNA contacts. It has been previously shown that short ssRNA oligonucleotides can bind duplex DNA in a sequence-specific manner by Hoogsteen or reverse-Hoogsteen pairing in polypurine/polypyrimidine tracts (17–19). Such binding provides an attractive basis for the interactions observed here. The obvious alternative would be the formation of an ‘R-loop’, in which the ssRNA locally invades the DNA duplex and displaces the same-sequence strand. These two possibilities could be distinguished by their differential sensitivities to RNase H, which will cleave R-loops but not triplex structures (20); however, our experimental system does not permit addition of RNase H after the ssRNA/dsDNA interaction has been established. Although the formation of R-loops is a suggested mechanism for lncRNA-dependent gene regulation (21,22), we note that R-loops normally form under conditions, which favors dsDNA denaturation, which is not the case in the current study. In either case, longer ssRNAs are predicted to exhibit stronger effects on dsDNA extension because they contain more local tracts suitable for interaction.
To further probe the plausibility of triplex formation as the basis for the observed interaction, we predicted the occurrence of triplex target sites (TTSs) along λ dsDNA using an algorithm termed Triplexator (23). As shown in Supplementary Table S2, when the length of the target sequence is set at a value larger than 19 bp, only two hits (22 793–22 814, 40 465–40 487) are obtained. As it has been reported that 12-nt of triplex-forming oligonucleotide is sufficient to form the triple helix even at neutral pH (24), we set the length threshold of TTS as 10 nt. 6L and 6H obtain 11 and 4 hits, respectively. The second longest TTS (20-nt) of λ dsDNA is found in 6L (Supplementary Table S2, Figure 1B). This might in part explain why the effect of 6L is more distinct than 6H. The five ssRNAs in Figure 3C: 40, 130, 416, 1025 and 6396 exhibit 1, 1, 2, 2 and 11 hits, respectively. Since the proposed interspersed local interaction requires at least two RNA/DNA bindings, the failure of 40 and 130-nt ssRNAs to give a detectable signal in force–extension analysis might be attributable to the presence of only a single possible interaction site. Alternatively, or in addition, the polypurine/polypyrimidine tracts in these ssRNAs may be sequestered in ssRNA secondary structure, whose formation will compete with the formation of RNA/DNA interactions.
Finally, our findings suggest interesting new possibilities for the effects of lncRNAs in vivo. The effects observed here specifically pertain to long ssRNAs, ≥1 kb. By implication, in vivo, the observed effects will pertain to lncRNAs that are relatively long. Most importantly, the presented findings suggest that a sufficiently long ncRNA has the potential to interact with two or more different homologous regions. By our proposed hypothesis, such interactions could bring two or more regions along the same chromosome into closer proximity, thus favoring other interactions in the affected region. Alternatively or in addition, such interactions could juxtapose two or more regions on different chromosomes, which might be homologs or non-homologous chromosomes that happen to share local homology.
Our finding also implies that periodic lncRNA/dsDNA interactions will alter the state of the dsDNA partner out of its most natural conformation. In addition to increasing the proximities of interacting regions, such effects might alter dsDNA supercoiling status; thereby, destabilizing nucleosomes or promoting protein binding.
Overall, ssRNA/dsDNA interactions could create complex two- and three-dimensional meshworks comprising ssRNA and altered-conformation dsDNA segments.
In summary, the presented results show that it is possible for long ssRNAs to interact directly with an homologous dsDNA, and that such interactions have the potential to alter the mechanical properties (and, by our hypothesis, the conformation) of the dsDNA. These findings provide emerging evidence that the in vivo roles of lncRNAs involve such direct interactions. RNA/dsDNA triplex interactions identified thus far in vivo involve single contact points between the two molecules (8,10,11). The present observations suggest that more complex interactions should also be possible. It also seems likely that the detected contacts involve paranemic interactions of the two molecules. A long ssRNA is unlikely to wind plectonemically around the dsDNA, which is tethered at both ends. Analogous interactions between ssRNA and dsDNA in vivo would lead to a situation in which a lncRNA is periodically associated with its chromosomal target but with a complex array of sequence-specific primary and secondary ssRNA structures in the intervening regions that would be available for interaction with proteins (or small RNAs) as needed for formation of specific chromatin structures and/or networks.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
National Institutes of Health [RO1 GM025326 to N.K.]. Funding for open access charge National Institutes of Health [R01 GM025326]; Shenzhen grants [JCYJ20140610152828703 and CXZZ20140901004122088 to C.L.].
Conflict of interest statement. None declared.




Comments